David Marlevi
Potential and challenges of generative adversarial networks for super-resolution in 4D Flow MRI
Aug 20, 2025Abstract:4D Flow Magnetic Resonance Imaging (4D Flow MRI) enables non-invasive quantification of blood flow and hemodynamic parameters. However, its clinical application is limited by low spatial resolution and noise, particularly affecting near-wall velocity measurements. Machine learning-based super-resolution has shown promise in addressing these limitations, but challenges remain, not least in recovering near-wall velocities. Generative adversarial networks (GANs) offer a compelling solution, having demonstrated strong capabilities in restoring sharp boundaries in non-medical super-resolution tasks. Yet, their application in 4D Flow MRI remains unexplored, with implementation challenged by known issues such as training instability and non-convergence. In this study, we investigate GAN-based super-resolution in 4D Flow MRI. Training and validation were conducted using patient-specific cerebrovascular in-silico models, converted into synthetic images via an MR-true reconstruction pipeline. A dedicated GAN architecture was implemented and evaluated across three adversarial loss functions: Vanilla, Relativistic, and Wasserstein. Our results demonstrate that the proposed GAN improved near-wall velocity recovery compared to a non-adversarial reference (vNRMSE: 6.9% vs. 9.6%); however, that implementation specifics are critical for stable network training. While Vanilla and Relativistic GANs proved unstable compared to generator-only training (vNRMSE: 8.1% and 7.8% vs. 7.2%), a Wasserstein GAN demonstrated optimal stability and incremental improvement (vNRMSE: 6.9% vs. 7.2%). The Wasserstein GAN further outperformed the generator-only baseline at low SNR (vNRMSE: 8.7% vs. 10.7%). These findings highlight the potential of GAN-based super-resolution in enhancing 4D Flow MRI, particularly in challenging cerebrovascular regions, while emphasizing the need for careful selection of adversarial strategies.
A Computational Pipeline for Advanced Analysis of 4D Flow MRI in the Left Atrium
May 14, 2025Abstract:The left atrium (LA) plays a pivotal role in modulating left ventricular filling, but our comprehension of its hemodynamics is significantly limited by the constraints of conventional ultrasound analysis. 4D flow magnetic resonance imaging (4D Flow MRI) holds promise for enhancing our understanding of atrial hemodynamics. However, the low velocities within the LA and the limited spatial resolution of 4D Flow MRI make analyzing this chamber challenging. Furthermore, the absence of dedicated computational frameworks, combined with diverse acquisition protocols and vendors, complicates gathering large cohorts for studying the prognostic value of hemodynamic parameters provided by 4D Flow MRI. In this study, we introduce the first open-source computational framework tailored for the analysis of 4D Flow MRI in the LA, enabling comprehensive qualitative and quantitative analysis of advanced hemodynamic parameters. Our framework proves robust to data from different centers of varying quality, producing high-accuracy automated segmentations (Dice $>$ 0.9 and Hausdorff 95 $<$ 3 mm), even with limited training data. Additionally, we conducted the first comprehensive assessment of energy, vorticity, and pressure parameters in the LA across a spectrum of disorders to investigate their potential as prognostic biomarkers.
Deep learning for temporal super-resolution 4D Flow MRI
Jan 15, 2025Abstract:4D Flow Magnetic Resonance Imaging (4D Flow MRI) is a non-invasive technique for volumetric, time-resolved blood flow quantification. However, apparent trade-offs between acquisition time, image noise, and resolution limit clinical applicability. In particular, in regions of highly transient flow, coarse temporal resolution can hinder accurate capture of physiologically relevant flow variations. To overcome these issues, post-processing techniques using deep learning have shown promising results to enhance resolution post-scan using so-called super-resolution networks. However, while super-resolution has been focusing on spatial upsampling, temporal super-resolution remains largely unexplored. The aim of this study was therefore to implement and evaluate a residual network for temporal super-resolution 4D Flow MRI. To achieve this, an existing spatial network (4DFlowNet) was re-designed for temporal upsampling, adapting input dimensions, and optimizing internal layer structures. Training and testing were performed using synthetic 4D Flow MRI data originating from patient-specific in-silico models, as well as using in-vivo datasets. Overall, excellent performance was achieved with input velocities effectively denoised and temporally upsampled, with a mean absolute error (MAE) of 1.0 cm/s in an unseen in-silico setting, outperforming deterministic alternatives (linear interpolation MAE = 2.3 cm/s, sinc interpolation MAE = 2.6 cm/s). Further, the network synthesized high-resolution temporal information from unseen low-resolution in-vivo data, with strong correlation observed at peak flow frames. As such, our results highlight the potential of utilizing data-driven neural networks for temporal super-resolution 4D Flow MRI, enabling high-frame-rate flow quantification without extending acquisition times beyond clinically acceptable limits.
Opportunities for machine learning in scientific discovery
May 07, 2024Abstract:Technological advancements have substantially increased computational power and data availability, enabling the application of powerful machine-learning (ML) techniques across various fields. However, our ability to leverage ML methods for scientific discovery, {\it i.e.} to obtain fundamental and formalized knowledge about natural processes, is still in its infancy. In this review, we explore how the scientific community can increasingly leverage ML techniques to achieve scientific discoveries. We observe that the applicability and opportunity of ML depends strongly on the nature of the problem domain, and whether we have full ({\it e.g.}, turbulence), partial ({\it e.g.}, computational biochemistry), or no ({\it e.g.}, neuroscience) {\it a-priori} knowledge about the governing equations and physical properties of the system. Although challenges remain, principled use of ML is opening up new avenues for fundamental scientific discoveries. Throughout these diverse fields, there is a theme that ML is enabling researchers to embrace complexity in observational data that was previously intractable to classic analysis and numerical investigations.
Generalized super-resolution 4D Flow MRI $\unicode{x2013}$ using ensemble learning to extend across the cardiovascular system
Nov 21, 2023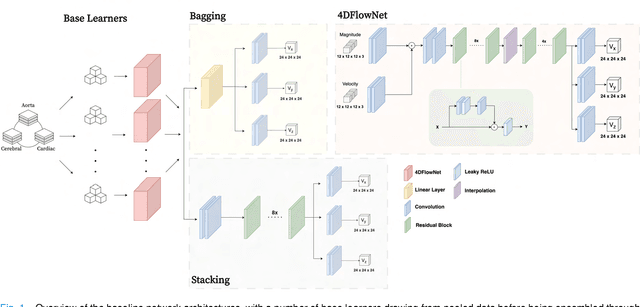
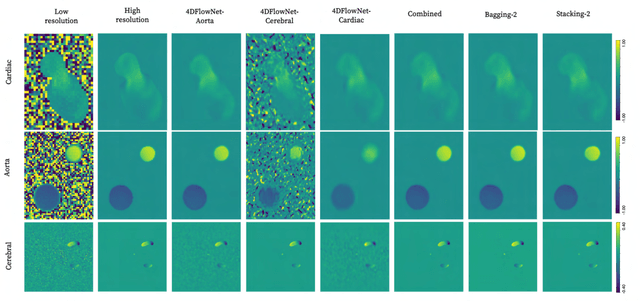
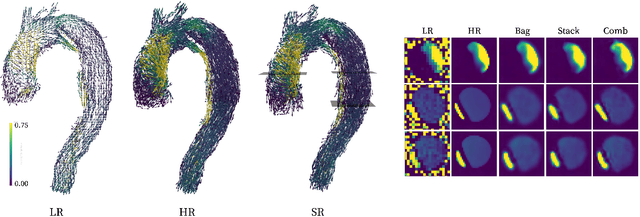
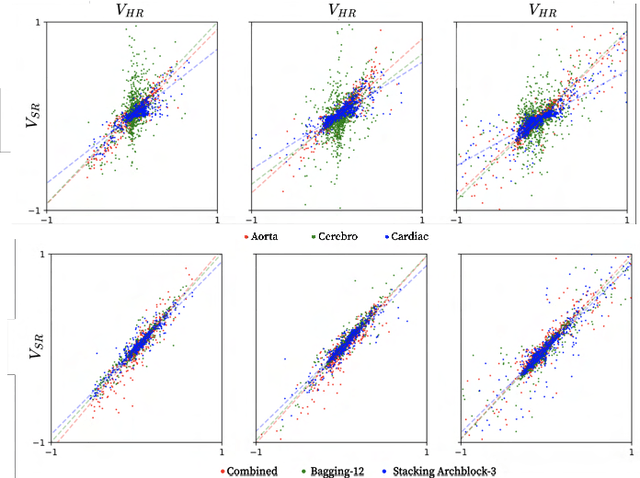
Abstract:4D Flow Magnetic Resonance Imaging (4D Flow MRI) is a non-invasive measurement technique capable of quantifying blood flow across the cardiovascular system. While practical use is limited by spatial resolution and image noise, incorporation of trained super-resolution (SR) networks has potential to enhance image quality post-scan. However, these efforts have predominantly been restricted to narrowly defined cardiovascular domains, with limited exploration of how SR performance extends across the cardiovascular system; a task aggravated by contrasting hemodynamic conditions apparent across the cardiovasculature. The aim of our study was to explore the generalizability of SR 4D Flow MRI using a combination of heterogeneous training sets and dedicated ensemble learning. With synthetic training data generated across three disparate domains (cardiac, aortic, cerebrovascular), varying convolutional base and ensemble learners were evaluated as a function of domain and architecture, quantifying performance on both in-silico and acquired in-vivo data from the same three domains. Results show that both bagging and stacking ensembling enhance SR performance across domains, accurately predicting high-resolution velocities from low-resolution input data in-silico. Likewise, optimized networks successfully recover native resolution velocities from downsampled in-vivo data, as well as show qualitative potential in generating denoised SR-images from clinical level input data. In conclusion, our work presents a viable approach for generalized SR 4D Flow MRI, with ensemble learning extending utility across various clinical areas of interest.
Morphology-based non-rigid registration of coronary computed tomography and intravascular images through virtual catheter path optimization
Dec 30, 2022



Abstract:Coronary Computed Tomography Angiography (CCTA) provides information on the presence, extent, and severity of obstructive coronary artery disease. Large-scale clinical studies analyzing CCTA-derived metrics typically require ground-truth validation in the form of high-fidelity 3D intravascular imaging. However, manual rigid alignment of intravascular images to corresponding CCTA images is both time consuming and user-dependent. Moreover, intravascular modalities suffer from several non-rigid motion-induced distortions arising from distortions in the imaging catheter path. To address these issues, we here present a semi-automatic segmentation-based framework for both rigid and non-rigid matching of intravascular images to CCTA images. We formulate the problem in terms of finding the optimal \emph{virtual catheter path} that samples the CCTA data to recapitulate the coronary artery morphology found in the intravascular image. We validate our co-registration framework on a cohort of $n=40$ patients using bifurcation landmarks as ground truth for longitudinal and rotational registration. Our results indicate that our non-rigid registration significantly outperforms other co-registration approaches for luminal bifurcation alignment in both longitudinal (mean mismatch: 3.3 frames) and rotational directions (mean mismatch: 28.6 degrees). By providing a differentiable framework for automatic multi-modal intravascular data fusion, our developed co-registration modules significantly reduces the manual effort required to conduct large-scale multi-modal clinical studies while also providing a solid foundation for the development of machine learning-based co-registration approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge