Muhammad Usman Akbar
Generalized super-resolution 4D Flow MRI $\unicode{x2013}$ using ensemble learning to extend across the cardiovascular system
Nov 21, 2023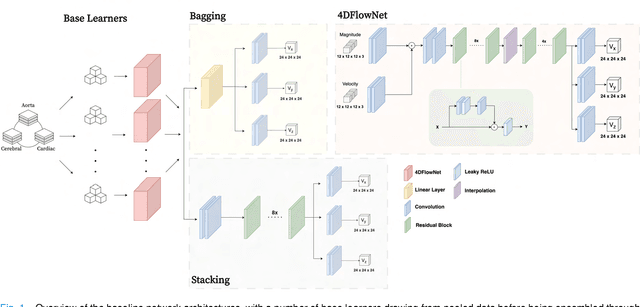
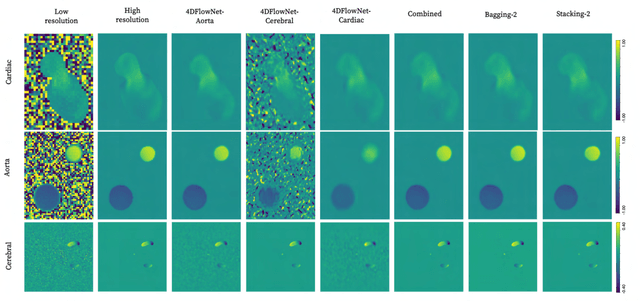
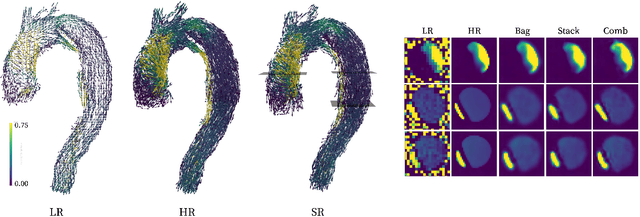
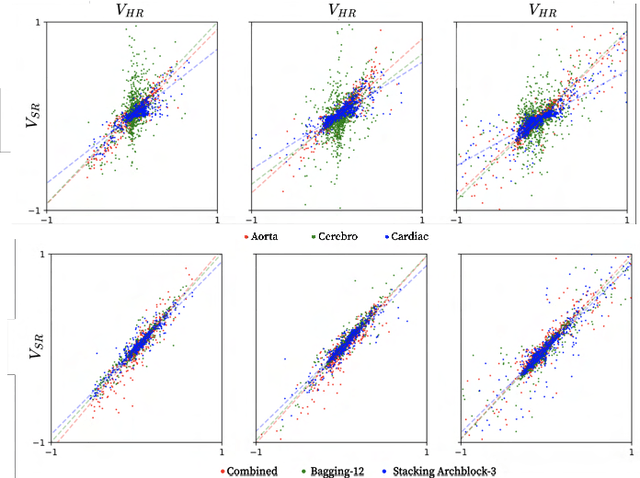
Abstract:4D Flow Magnetic Resonance Imaging (4D Flow MRI) is a non-invasive measurement technique capable of quantifying blood flow across the cardiovascular system. While practical use is limited by spatial resolution and image noise, incorporation of trained super-resolution (SR) networks has potential to enhance image quality post-scan. However, these efforts have predominantly been restricted to narrowly defined cardiovascular domains, with limited exploration of how SR performance extends across the cardiovascular system; a task aggravated by contrasting hemodynamic conditions apparent across the cardiovasculature. The aim of our study was to explore the generalizability of SR 4D Flow MRI using a combination of heterogeneous training sets and dedicated ensemble learning. With synthetic training data generated across three disparate domains (cardiac, aortic, cerebrovascular), varying convolutional base and ensemble learners were evaluated as a function of domain and architecture, quantifying performance on both in-silico and acquired in-vivo data from the same three domains. Results show that both bagging and stacking ensembling enhance SR performance across domains, accurately predicting high-resolution velocities from low-resolution input data in-silico. Likewise, optimized networks successfully recover native resolution velocities from downsampled in-vivo data, as well as show qualitative potential in generating denoised SR-images from clinical level input data. In conclusion, our work presents a viable approach for generalized SR 4D Flow MRI, with ensemble learning extending utility across various clinical areas of interest.
Brain tumor segmentation using synthetic MR images -- A comparison of GANs and diffusion models
Jun 05, 2023Abstract:Large annotated datasets are required for training deep learning models, but in medical imaging data sharing is often complicated due to ethics, anonymization and data protection legislation (e.g. the general data protection regulation (GDPR)). Generative AI models, such as generative adversarial networks (GANs) and diffusion models, can today produce very realistic synthetic images, and can potentially facilitate data sharing as GDPR should not apply for medical images which do not belong to a specific person. However, in order to share synthetic images it must first be demonstrated that they can be used for training different networks with acceptable performance. Here, we therefore comprehensively evaluate four GANs (progressive GAN, StyleGAN 1-3) and a diffusion model for the task of brain tumor segmentation. Our results show that segmentation networks trained on synthetic images reach Dice scores that are 80\% - 90\% of Dice scores when training with real images, but that memorization of the training images can be a problem for diffusion models if the original dataset is too small. Furthermore, we demonstrate that common metrics for evaluating synthetic images, Fr\'echet inception distance (FID) and inception score (IS), do not correlate well with the obtained performance when using the synthetic images for training segmentation networks.
Beware of diffusion models for synthesizing medical images -- A comparison with GANs in terms of memorizing brain tumor images
May 12, 2023Abstract:Diffusion models were initially developed for text-to-image generation and are now being utilized to generate high quality synthetic images. Preceded by GANs, diffusion models have shown impressive results using various evaluation metrics. However, commonly used metrics such as FID and IS are not suitable for determining whether diffusion models are simply reproducing the training images. Here we train StyleGAN and diffusion models, using BRATS20 and BRATS21 datasets, to synthesize brain tumor images, and measure the correlation between the synthetic images and all training images. Our results show that diffusion models are much more likely to memorize the training images, especially for small datasets. Researchers should be careful when using diffusion models for medical imaging, if the final goal is to share the synthetic images.
Efficient brain age prediction from 3D MRI volumes using 2D projections
Nov 10, 2022Abstract:Using 3D CNNs on high resolution medical volumes is very computationally demanding, especially for large datasets like the UK Biobank which aims to scan 100,000 subjects. Here we demonstrate that using 2D CNNs on a few 2D projections (representing mean and standard deviation across axial, sagittal and coronal slices) of the 3D volumes leads to reasonable test accuracy when predicting the age from brain volumes. Using our approach, one training epoch with 20,324 subjects takes 40 - 70 seconds using a single GPU, which is almost 100 times faster compared to a small 3D CNN. These results are important for researchers who do not have access to expensive GPU hardware for 3D CNNs.
Does an ensemble of GANs lead to better performance when training segmentation networks with synthetic images?
Nov 08, 2022Abstract:Large annotated datasets are required to train segmentation networks. In medical imaging, it is often difficult, time consuming and expensive to create such datasets, and it may also be difficult to share these datasets with other researchers. Different AI models can today generate very realistic synthetic images, which can potentially be openly shared as they do not belong to specific persons. However, recent work has shown that using synthetic images for training deep networks often leads to worse performance compared to using real images. Here we demonstrate that using synthetic images and annotations from an ensemble of 10 GANs, instead of from a single GAN, increases the Dice score on real test images with 4.7 % to 14.0 % on specific classes.
AIforCOVID: predicting the clinical outcomes in patients with COVID-19 applying AI to chest-X-rays. An Italian multicentre study
Dec 11, 2020
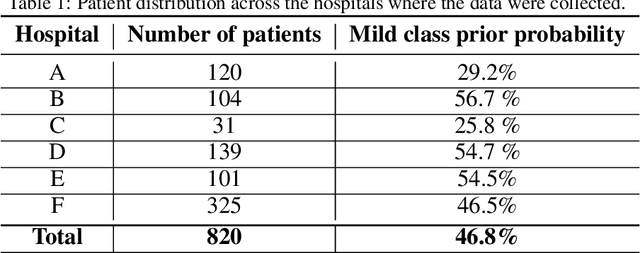
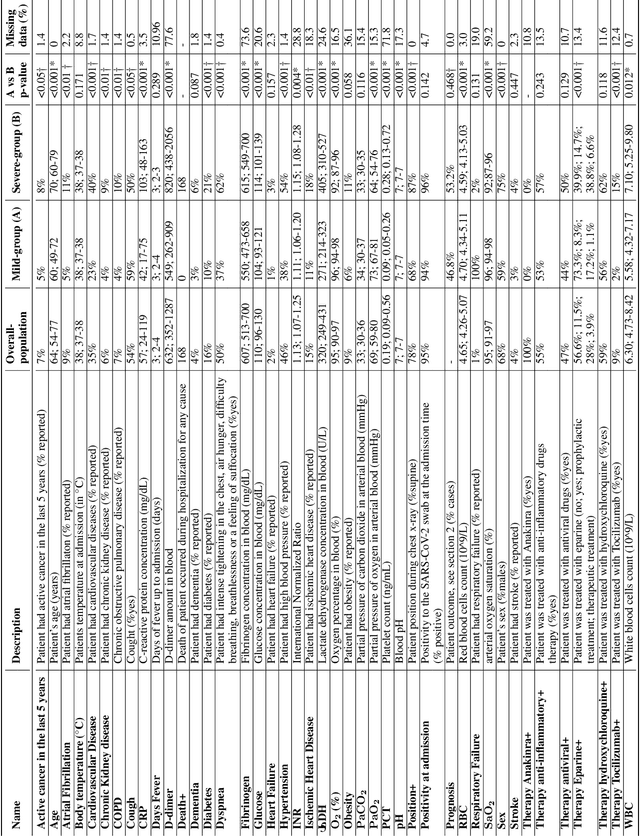
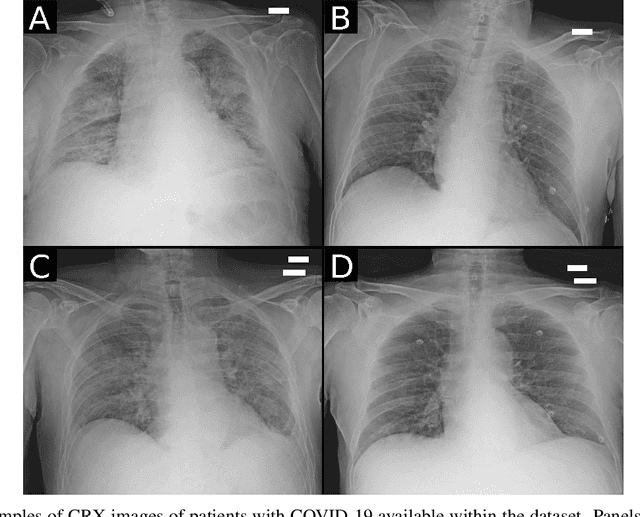
Abstract:Recent epidemiological data report that worldwide more than 53 million people have been infected by SARS-CoV-2, resulting in 1.3 million deaths. The disease has been spreading very rapidly and few months after the identification of the first infected, shortage of hospital resources quickly became a problem. In this work we investigate whether chest X-ray (CXR) can be used as a possible tool for the early identification of patients at risk of severe outcome, like intensive care or death. CXR is a radiological technique that compared to computed tomography (CT) it is simpler, faster, more widespread and it induces lower radiation dose. We present a dataset including data collected from 820 patients by six Italian hospitals in spring 2020 during the first COVID-19 emergency. The dataset includes CXR images, several clinical attributes and clinical outcomes. We investigate the potential of artificial intelligence to predict the prognosis of such patients, distinguishing between severe and mild cases, thus offering a baseline reference for other researchers and practitioners. To this goal, we present three approaches that use features extracted from CXR images, either handcrafted or automatically by convolutional neuronal networks, which are then integrated with the clinical data. Exhaustive evaluation shows promising performance both in 10-fold and leave-one-centre-out cross-validation, implying that clinical data and images have the potential to provide useful information for the management of patients and hospital resources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge