David Nordsletten
Deep learning for temporal super-resolution 4D Flow MRI
Jan 15, 2025Abstract:4D Flow Magnetic Resonance Imaging (4D Flow MRI) is a non-invasive technique for volumetric, time-resolved blood flow quantification. However, apparent trade-offs between acquisition time, image noise, and resolution limit clinical applicability. In particular, in regions of highly transient flow, coarse temporal resolution can hinder accurate capture of physiologically relevant flow variations. To overcome these issues, post-processing techniques using deep learning have shown promising results to enhance resolution post-scan using so-called super-resolution networks. However, while super-resolution has been focusing on spatial upsampling, temporal super-resolution remains largely unexplored. The aim of this study was therefore to implement and evaluate a residual network for temporal super-resolution 4D Flow MRI. To achieve this, an existing spatial network (4DFlowNet) was re-designed for temporal upsampling, adapting input dimensions, and optimizing internal layer structures. Training and testing were performed using synthetic 4D Flow MRI data originating from patient-specific in-silico models, as well as using in-vivo datasets. Overall, excellent performance was achieved with input velocities effectively denoised and temporally upsampled, with a mean absolute error (MAE) of 1.0 cm/s in an unseen in-silico setting, outperforming deterministic alternatives (linear interpolation MAE = 2.3 cm/s, sinc interpolation MAE = 2.6 cm/s). Further, the network synthesized high-resolution temporal information from unseen low-resolution in-vivo data, with strong correlation observed at peak flow frames. As such, our results highlight the potential of utilizing data-driven neural networks for temporal super-resolution 4D Flow MRI, enabling high-frame-rate flow quantification without extending acquisition times beyond clinically acceptable limits.
Generalized super-resolution 4D Flow MRI $\unicode{x2013}$ using ensemble learning to extend across the cardiovascular system
Nov 21, 2023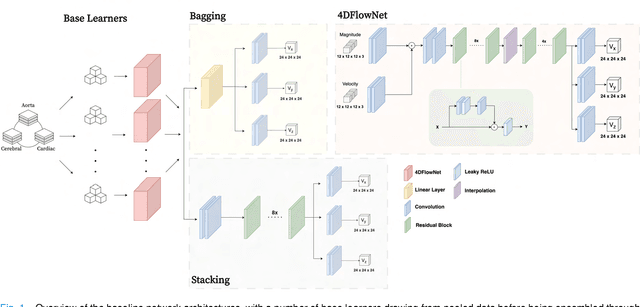
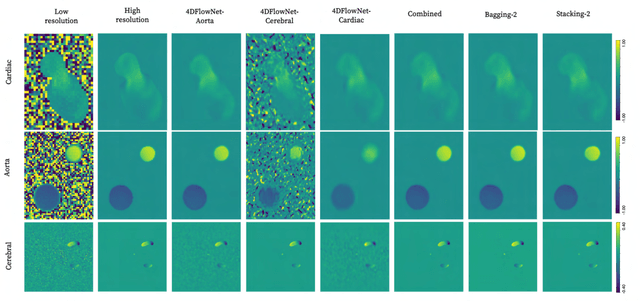
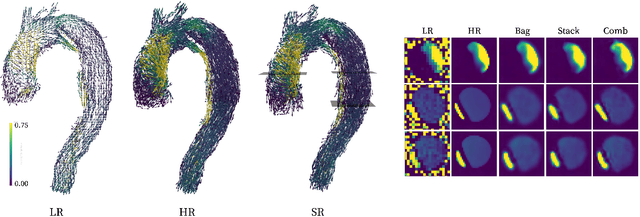
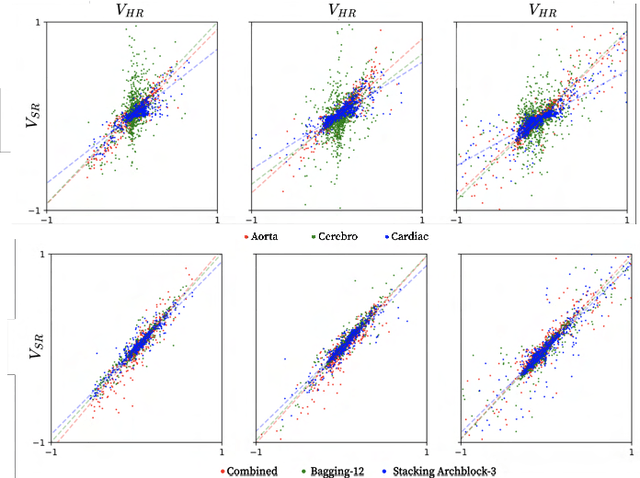
Abstract:4D Flow Magnetic Resonance Imaging (4D Flow MRI) is a non-invasive measurement technique capable of quantifying blood flow across the cardiovascular system. While practical use is limited by spatial resolution and image noise, incorporation of trained super-resolution (SR) networks has potential to enhance image quality post-scan. However, these efforts have predominantly been restricted to narrowly defined cardiovascular domains, with limited exploration of how SR performance extends across the cardiovascular system; a task aggravated by contrasting hemodynamic conditions apparent across the cardiovasculature. The aim of our study was to explore the generalizability of SR 4D Flow MRI using a combination of heterogeneous training sets and dedicated ensemble learning. With synthetic training data generated across three disparate domains (cardiac, aortic, cerebrovascular), varying convolutional base and ensemble learners were evaluated as a function of domain and architecture, quantifying performance on both in-silico and acquired in-vivo data from the same three domains. Results show that both bagging and stacking ensembling enhance SR performance across domains, accurately predicting high-resolution velocities from low-resolution input data in-silico. Likewise, optimized networks successfully recover native resolution velocities from downsampled in-vivo data, as well as show qualitative potential in generating denoised SR-images from clinical level input data. In conclusion, our work presents a viable approach for generalized SR 4D Flow MRI, with ensemble learning extending utility across various clinical areas of interest.
Estimation of Cardiac Valve Annuli Motion with Deep Learning
Oct 23, 2020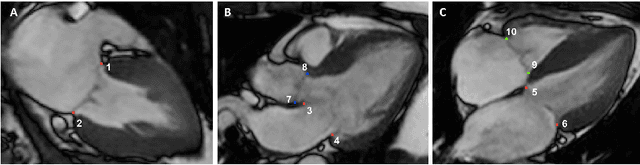


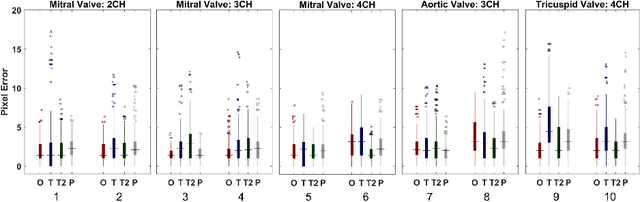
Abstract:Valve annuli motion and morphology, measured from non-invasive imaging, can be used to gain a better understanding of healthy and pathological heart function. Measurements such as long-axis strain as well as peak strain rates provide markers of systolic function. Likewise, early and late-diastolic filling velocities are used as indicators of diastolic function. Quantifying global strains, however, requires a fast and precise method of tracking long-axis motion throughout the cardiac cycle. Valve landmarks such as the insertion of leaflets into the myocardial wall provide features that can be tracked to measure global long-axis motion. Feature tracking methods require initialisation, which can be time-consuming in studies with large cohorts. Therefore, this study developed and trained a neural network to identify ten features from unlabeled long-axis MR images: six mitral valve points from three long-axis views, two aortic valve points and two tricuspid valve points. This study used manual annotations of valve landmarks in standard 2-, 3- and 4-chamber long-axis images collected in clinical scans to train the network. The accuracy in the identification of these ten features, in pixel distance, was compared with the accuracy of two commonly used feature tracking methods as well as the inter-observer variability of manual annotations. Clinical measures, such as valve landmark strain and motion between end-diastole and end-systole, are also presented to illustrate the utility and robustness of the method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge