David Stojanovski
DeepSPV: An Interpretable Deep Learning Pipeline for 3D Spleen Volume Estimation from 2D Ultrasound Images
Nov 17, 2024



Abstract:Splenomegaly, the enlargement of the spleen, is an important clinical indicator for various associated medical conditions, such as sickle cell disease (SCD). Spleen length measured from 2D ultrasound is the most widely used metric for characterising spleen size. However, it is still considered a surrogate measure, and spleen volume remains the gold standard for assessing spleen size. Accurate spleen volume measurement typically requires 3D imaging modalities, such as computed tomography or magnetic resonance imaging, but these are not widely available, especially in the Global South which has a high prevalence of SCD. In this work, we introduce a deep learning pipeline, DeepSPV, for precise spleen volume estimation from single or dual 2D ultrasound images. The pipeline involves a segmentation network and a variational autoencoder for learning low-dimensional representations from the estimated segmentations. We investigate three approaches for spleen volume estimation and our best model achieves 86.62%/92.5% mean relative volume accuracy (MRVA) under single-view/dual-view settings, surpassing the performance of human experts. In addition, the pipeline can provide confidence intervals for the volume estimates as well as offering benefits in terms of interpretability, which further support clinicians in decision-making when identifying splenomegaly. We evaluate the full pipeline using a highly realistic synthetic dataset generated by a diffusion model, achieving an overall MRVA of 83.0% from a single 2D ultrasound image. Our proposed DeepSPV is the first work to use deep learning to estimate 3D spleen volume from 2D ultrasound images and can be seamlessly integrated into the current clinical workflow for spleen assessment.
Efficient Semantic Diffusion Architectures for Model Training on Synthetic Echocardiograms
Sep 28, 2024Abstract:We investigate the utility of diffusion generative models to efficiently synthesise datasets that effectively train deep learning models for image analysis. Specifically, we propose novel $\Gamma$-distribution Latent Denoising Diffusion Models (LDMs) designed to generate semantically guided synthetic cardiac ultrasound images with improved computational efficiency. We also investigate the potential of using these synthetic images as a replacement for real data in training deep networks for left-ventricular segmentation and binary echocardiogram view classification tasks. We compared six diffusion models in terms of the computational cost of generating synthetic 2D echo data, the visual realism of the resulting images, and the performance, on real data, of downstream tasks (segmentation and classification) trained using these synthetic echoes. We compare various diffusion strategies and ODE solvers for their impact on segmentation and classification performance. The results show that our propose architectures significantly reduce computational costs while maintaining or improving downstream task performance compared to state-of-the-art methods. While other diffusion models generated more realistic-looking echo images at higher computational cost, our research suggests that for model training, visual realism is not necessarily related to model performance, and considerable compute costs can be saved by using more efficient models.
Echo from noise: synthetic ultrasound image generation using diffusion models for real image segmentation
May 09, 2023Abstract:We propose a novel pipeline for the generation of synthetic images via Denoising Diffusion Probabilistic Models (DDPMs) guided by cardiac ultrasound semantic label maps. We show that these synthetic images can serve as a viable substitute for real data in the training of deep-learning models for medical image analysis tasks such as image segmentation. To demonstrate the effectiveness of this approach, we generated synthetic 2D echocardiography images and trained a neural network for segmentation of the left ventricle and left atrium. The performance of the network trained on exclusively synthetic images was evaluated on an unseen dataset of real images and yielded mean Dice scores of 88.5 $\pm 6.0$ , 92.3 $\pm 3.9$, 86.3 $\pm 10.7$ \% for left ventricular endocardial, epicardial and left atrial segmentation respectively. This represents an increase of $9.09$, $3.7$ and $15.0$ \% in Dice scores compared to the previous state-of-the-art. The proposed pipeline has the potential for application to a wide range of other tasks across various medical imaging modalities.
Efficient Pix2Vox++ for 3D Cardiac Reconstruction from 2D echo views
Jul 27, 2022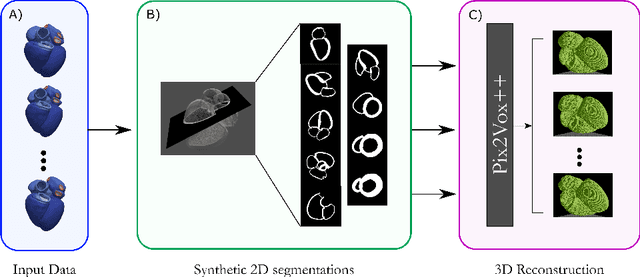
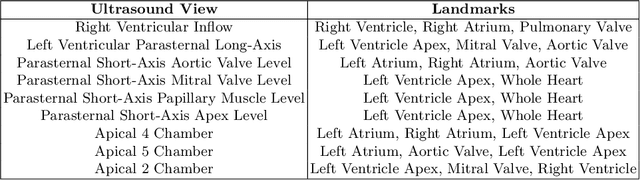
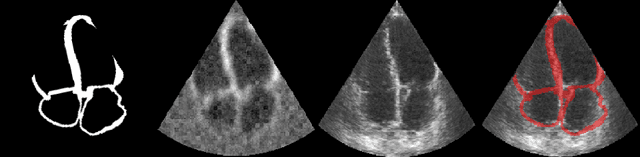
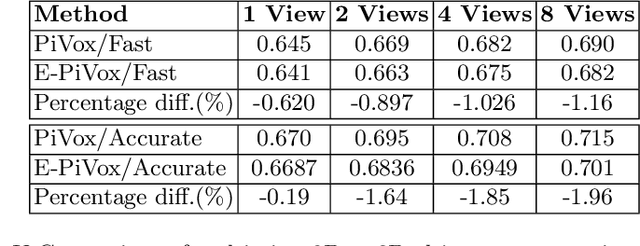
Abstract:Accurate geometric quantification of the human heart is a key step in the diagnosis of numerous cardiac diseases, and in the management of cardiac patients. Ultrasound imaging is the primary modality for cardiac imaging, however acquisition requires high operator skill, and its interpretation and analysis is difficult due to artifacts. Reconstructing cardiac anatomy in 3D can enable discovery of new biomarkers and make imaging less dependent on operator expertise, however most ultrasound systems only have 2D imaging capabilities. We propose both a simple alteration to the Pix2Vox++ networks for a sizeable reduction in memory usage and computational complexity, and a pipeline to perform reconstruction of 3D anatomy from 2D standard cardiac views, effectively enabling 3D anatomical reconstruction from limited 2D data. We evaluate our pipeline using synthetically generated data achieving accurate 3D whole-heart reconstructions (peak intersection over union score > 0.88) from just two standard anatomical 2D views of the heart. We also show preliminary results using real echo images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge