Pablo Lamata
Cardiac Digital Twins at Scale from MRI: Open Tools and Representative Models from ~55000 UK Biobank Participants
May 27, 2025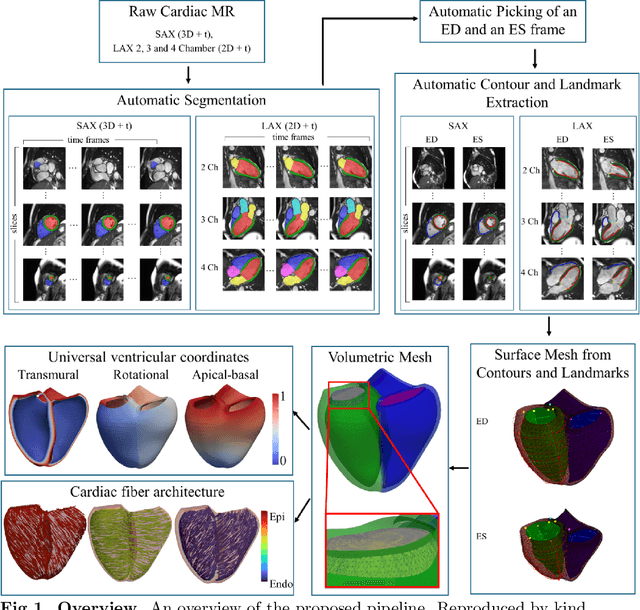
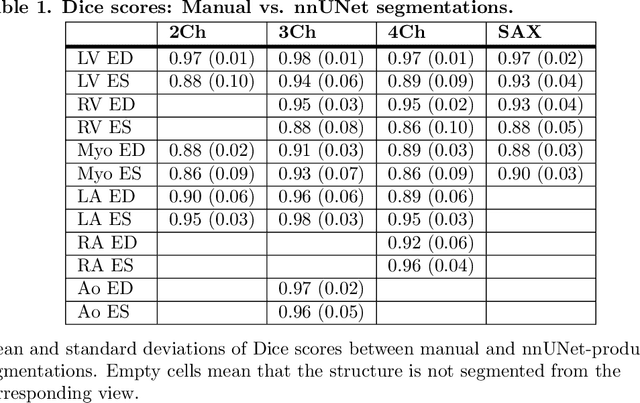
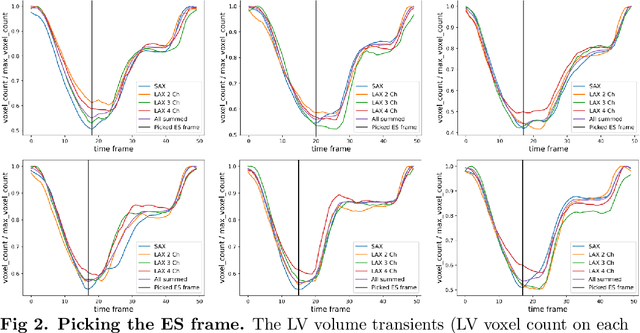
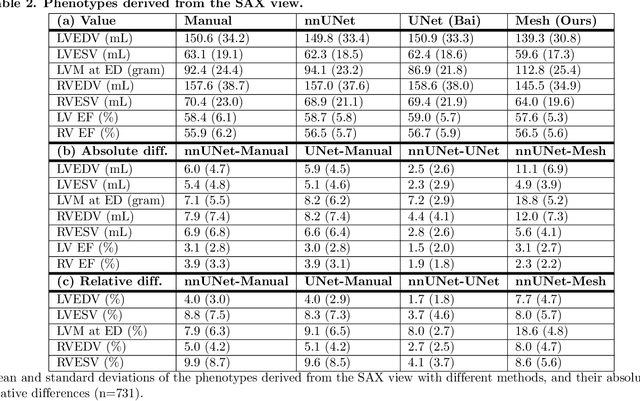
Abstract:A cardiac digital twin is a virtual replica of a patient's heart for screening, diagnosis, prognosis, risk assessment, and treatment planning of cardiovascular diseases. This requires an anatomically accurate patient-specific 3D structural representation of the heart, suitable for electro-mechanical simulations or study of disease mechanisms. However, generation of cardiac digital twins at scale is demanding and there are no public repositories of models across demographic groups. We describe an automatic open-source pipeline for creating patient-specific left and right ventricular meshes from cardiovascular magnetic resonance images, its application to a large cohort of ~55000 participants from UK Biobank, and the construction of the most comprehensive cohort of adult heart models to date, comprising 1423 representative meshes across sex (male, female), body mass index (range: 16 - 42 kg/m$^2$) and age (range: 49 - 80 years). Our code is available at https://github.com/cdttk/biv-volumetric-meshing/tree/plos2025 , and pre-trained networks, representative volumetric meshes with fibers and UVCs will be made available soon.
Efficient Semantic Diffusion Architectures for Model Training on Synthetic Echocardiograms
Sep 28, 2024Abstract:We investigate the utility of diffusion generative models to efficiently synthesise datasets that effectively train deep learning models for image analysis. Specifically, we propose novel $\Gamma$-distribution Latent Denoising Diffusion Models (LDMs) designed to generate semantically guided synthetic cardiac ultrasound images with improved computational efficiency. We also investigate the potential of using these synthetic images as a replacement for real data in training deep networks for left-ventricular segmentation and binary echocardiogram view classification tasks. We compared six diffusion models in terms of the computational cost of generating synthetic 2D echo data, the visual realism of the resulting images, and the performance, on real data, of downstream tasks (segmentation and classification) trained using these synthetic echoes. We compare various diffusion strategies and ODE solvers for their impact on segmentation and classification performance. The results show that our propose architectures significantly reduce computational costs while maintaining or improving downstream task performance compared to state-of-the-art methods. While other diffusion models generated more realistic-looking echo images at higher computational cost, our research suggests that for model training, visual realism is not necessarily related to model performance, and considerable compute costs can be saved by using more efficient models.
Echo from noise: synthetic ultrasound image generation using diffusion models for real image segmentation
May 09, 2023Abstract:We propose a novel pipeline for the generation of synthetic images via Denoising Diffusion Probabilistic Models (DDPMs) guided by cardiac ultrasound semantic label maps. We show that these synthetic images can serve as a viable substitute for real data in the training of deep-learning models for medical image analysis tasks such as image segmentation. To demonstrate the effectiveness of this approach, we generated synthetic 2D echocardiography images and trained a neural network for segmentation of the left ventricle and left atrium. The performance of the network trained on exclusively synthetic images was evaluated on an unseen dataset of real images and yielded mean Dice scores of 88.5 $\pm 6.0$ , 92.3 $\pm 3.9$, 86.3 $\pm 10.7$ \% for left ventricular endocardial, epicardial and left atrial segmentation respectively. This represents an increase of $9.09$, $3.7$ and $15.0$ \% in Dice scores compared to the previous state-of-the-art. The proposed pipeline has the potential for application to a wide range of other tasks across various medical imaging modalities.
Efficient Pix2Vox++ for 3D Cardiac Reconstruction from 2D echo views
Jul 27, 2022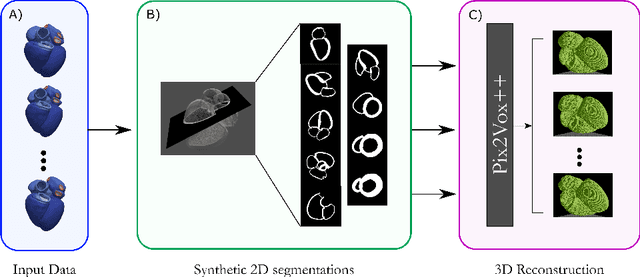
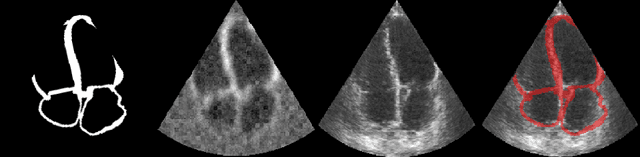
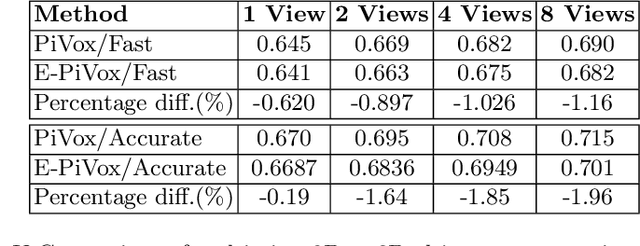
Abstract:Accurate geometric quantification of the human heart is a key step in the diagnosis of numerous cardiac diseases, and in the management of cardiac patients. Ultrasound imaging is the primary modality for cardiac imaging, however acquisition requires high operator skill, and its interpretation and analysis is difficult due to artifacts. Reconstructing cardiac anatomy in 3D can enable discovery of new biomarkers and make imaging less dependent on operator expertise, however most ultrasound systems only have 2D imaging capabilities. We propose both a simple alteration to the Pix2Vox++ networks for a sizeable reduction in memory usage and computational complexity, and a pipeline to perform reconstruction of 3D anatomy from 2D standard cardiac views, effectively enabling 3D anatomical reconstruction from limited 2D data. We evaluate our pipeline using synthetically generated data achieving accurate 3D whole-heart reconstructions (peak intersection over union score > 0.88) from just two standard anatomical 2D views of the heart. We also show preliminary results using real echo images.
A Global Benchmark of Algorithms for Segmenting Late Gadolinium-Enhanced Cardiac Magnetic Resonance Imaging
May 07, 2020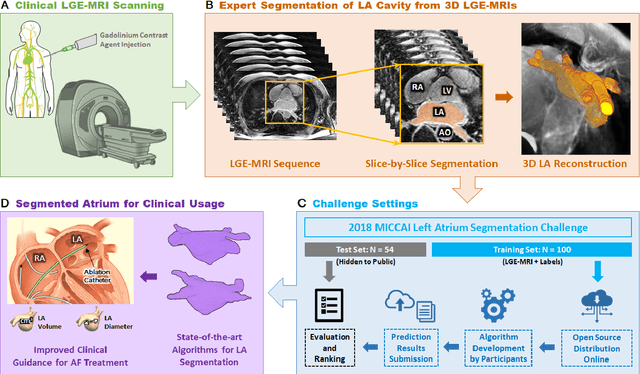
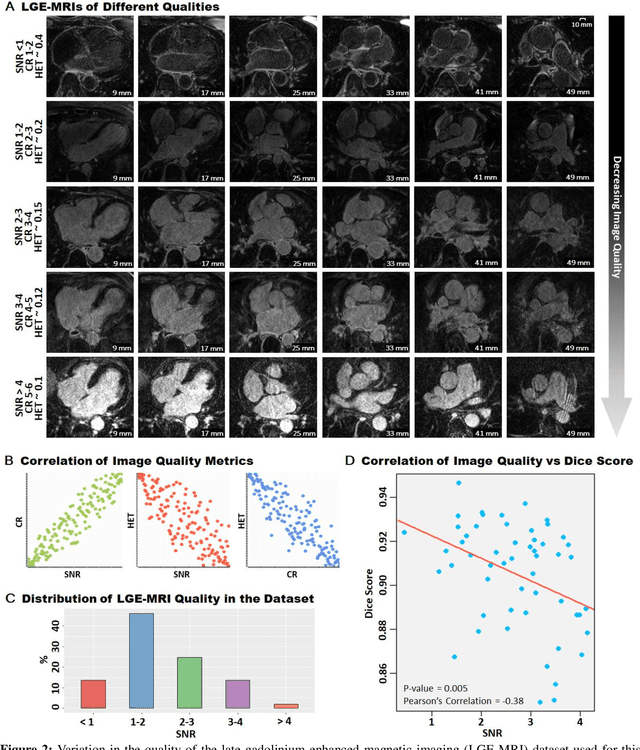
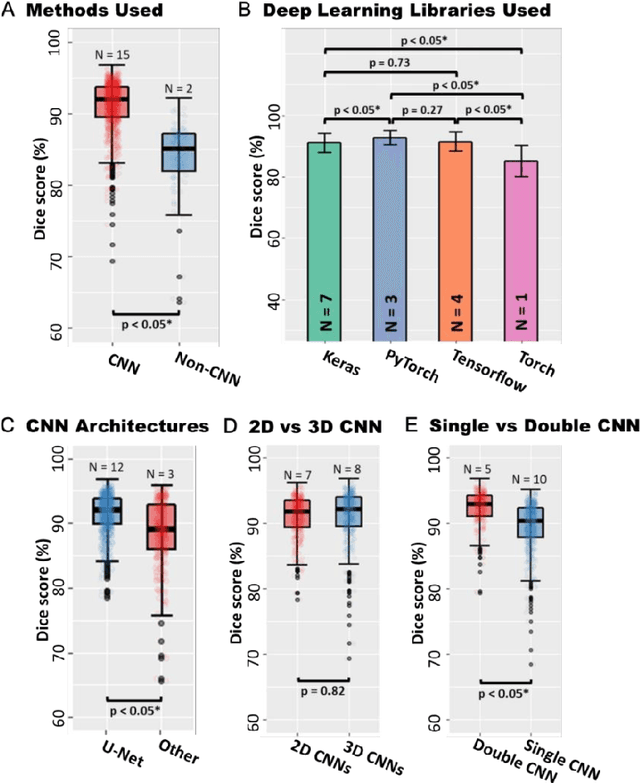
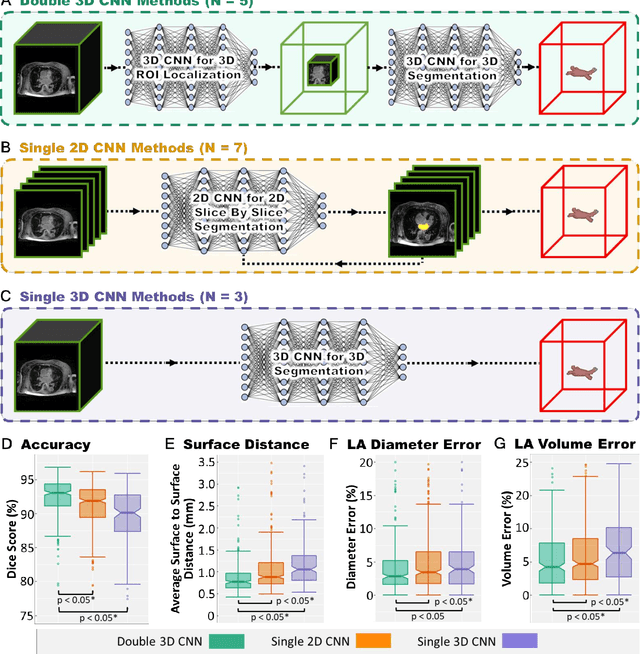
Abstract:Segmentation of cardiac images, particularly late gadolinium-enhanced magnetic resonance imaging (LGE-MRI) widely used for visualizing diseased cardiac structures, is a crucial first step for clinical diagnosis and treatment. However, direct segmentation of LGE-MRIs is challenging due to its attenuated contrast. Since most clinical studies have relied on manual and labor-intensive approaches, automatic methods are of high interest, particularly optimized machine learning approaches. To address this, we organized the "2018 Left Atrium Segmentation Challenge" using 154 3D LGE-MRIs, currently the world's largest cardiac LGE-MRI dataset, and associated labels of the left atrium segmented by three medical experts, ultimately attracting the participation of 27 international teams. In this paper, extensive analysis of the submitted algorithms using technical and biological metrics was performed by undergoing subgroup analysis and conducting hyper-parameter analysis, offering an overall picture of the major design choices of convolutional neural networks (CNNs) and practical considerations for achieving state-of-the-art left atrium segmentation. Results show the top method achieved a dice score of 93.2% and a mean surface to a surface distance of 0.7 mm, significantly outperforming prior state-of-the-art. Particularly, our analysis demonstrated that double, sequentially used CNNs, in which a first CNN is used for automatic region-of-interest localization and a subsequent CNN is used for refined regional segmentation, achieved far superior results than traditional methods and pipelines containing single CNNs. This large-scale benchmarking study makes a significant step towards much-improved segmentation methods for cardiac LGE-MRIs, and will serve as an important benchmark for evaluating and comparing the future works in the field.
A Generative Adversarial Model for Right Ventricle Segmentation
Sep 27, 2018
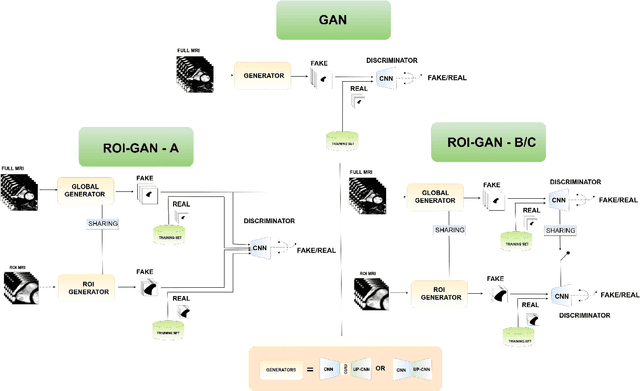
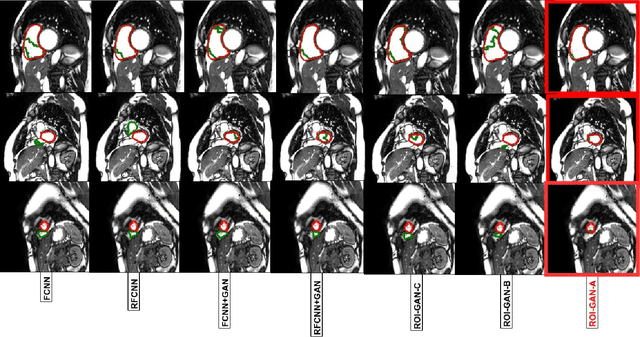

Abstract:The clinical management of several cardiovascular conditions, such as pulmonary hypertension, require the assessment of the right ventricular (RV) function. This work addresses the fully automatic and robust access to one of the key RV biomarkers, its ejection fraction, from the gold standard imaging modality, MRI. The problem becomes the accurate segmentation of the RV blood pool from cine MRI sequences. This work proposes a solution based on Fully Convolutional Neural Networks (FCNN), where our first contribution is the optimal combination of three concepts (the convolution Gated Recurrent Units (GRU), the Generative Adversarial Networks (GAN), and the L1 loss function) that achieves an improvement of 0.05 and 3.49 mm in Dice Index and Hausdorff Distance respectively with respect to the baseline FCNN. This improvement is then doubled by our second contribution, the ROI-GAN, that sets two GANs to cooperate working at two fields of view of the image, its full resolution and the region of interest (ROI). Our rationale here is to better guide the FCNN learning by combining global (full resolution) and local Region Of Interest (ROI) features. The study is conducted in a large in-house dataset of $\sim$ 23.000 segmented MRI slices, and its generality is verified in a publicly available dataset.
V-FCNN: Volumetric Fully Convolution Neural Network For Automatic Atrial Segmentation
Sep 27, 2018



Abstract:Atrial Fibrillation (AF) is a common electro-physiological cardiac disorder that causes changes in the anatomy of the atria. A better characterization of these changes is desirable for the definition of clinical biomarkers, furthermore, thus there is a need for its fully automatic segmentation from clinical images. In this work, we present an architecture based on 3D-convolution kernels, a Volumetric Fully Convolution Neural Network (V-FCNN), able to segment the entire volume in a one-shot, and consequently integrate the implicit spatial redundancy present in high-resolution images. A loss function based on the mixture of both Mean Square Error (MSE) and Dice Loss (DL) is used, in an attempt to combine the ability to capture the bulk shape as well as the reduction of local errors products by over-segmentation. Results demonstrate a reasonable performance in the middle region of the atria along with the impact of the challenges of capturing the variability of the pulmonary veins or the identification of the valve plane that separates the atria to the ventricle. A final dice of $92.5\%$ in $54$ patients ($4752$ atria test slices in total) is shown.
Automated segmentation on the entire cardiac cycle using a deep learning work-flow
Aug 31, 2018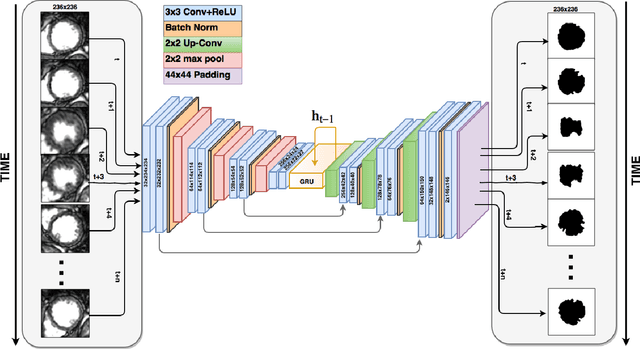
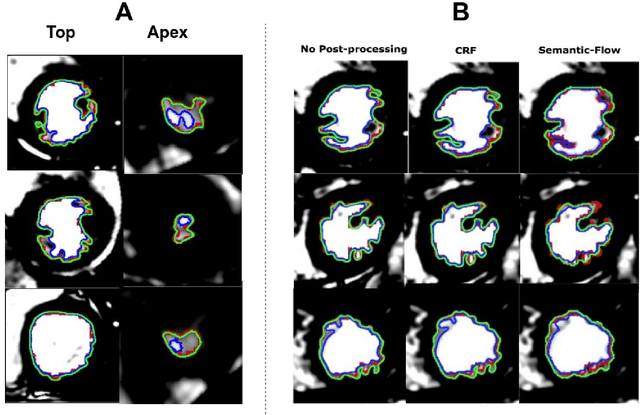
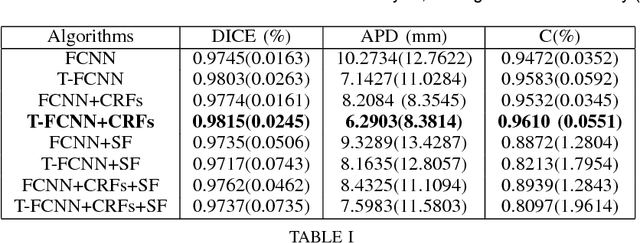
Abstract:The segmentation of the left ventricle (LV) from CINE MRI images is essential to infer important clinical parameters. Typically, machine learning algorithms for automated LV segmentation use annotated contours from only two cardiac phases, diastole, and systole. In this work, we present an analysis work-flow for fully-automated LV segmentation that learns from images acquired through the cardiac cycle. The workflow consists of three components: first, for each image in the sequence, we perform an automated localization and subsequent cropping of the bounding box containing the cardiac silhouette. Second, we identify the LV contours using a Temporal Fully Convolutional Neural Network (T-FCNN), which extends Fully Convolutional Neural Networks (FCNN) through a recurrent mechanism enforcing temporal coherence across consecutive frames. Finally, we further defined the boundaries using either one of two components: fully-connected Conditional Random Fields (CRFs) with Gaussian edge potentials and Semantic Flow. Our initial experiments suggest that significant improvement in performance can potentially be achieved by using a recurrent neural network component that explicitly learns cardiac motion patterns whilst performing LV segmentation.
Temporal Convolution Networks for Real-Time Abdominal Fetal Aorta Analysis with Ultrasound
Jul 11, 2018


Abstract:The automatic analysis of ultrasound sequences can substantially improve the efficiency of clinical diagnosis. In this work we present our attempt to automate the challenging task of measuring the vascular diameter of the fetal abdominal aorta from ultrasound images. We propose a neural network architecture consisting of three blocks: a convolutional layer for the extraction of imaging features, a Convolution Gated Recurrent Unit (C-GRU) for enforcing the temporal coherence across video frames and exploiting the temporal redundancy of a signal, and a regularized loss function, called \textit{CyclicLoss}, to impose our prior knowledge about the periodicity of the observed signal. We present experimental evidence suggesting that the proposed architecture can reach an accuracy substantially superior to previously proposed methods, providing an average reduction of the mean squared error from $0.31 mm^2$ (state-of-art) to $0.09 mm^2$, and a relative error reduction from $8.1\%$ to $5.3\%$. The mean execution speed of the proposed approach of 289 frames per second makes it suitable for real time clinical use.
Recurrent Fully Convolutional Neural Networks for Multi-slice MRI Cardiac Segmentation
Aug 13, 2016



Abstract:In cardiac magnetic resonance imaging, fully-automatic segmentation of the heart enables precise structural and functional measurements to be taken, e.g. from short-axis MR images of the left-ventricle. In this work we propose a recurrent fully-convolutional network (RFCN) that learns image representations from the full stack of 2D slices and has the ability to leverage inter-slice spatial dependences through internal memory units. RFCN combines anatomical detection and segmentation into a single architecture that is trained end-to-end thus significantly reducing computational time, simplifying the segmentation pipeline, and potentially enabling real-time applications. We report on an investigation of RFCN using two datasets, including the publicly available MICCAI 2009 Challenge dataset. Comparisons have been carried out between fully convolutional networks and deep restricted Boltzmann machines, including a recurrent version that leverages inter-slice spatial correlation. Our studies suggest that RFCN produces state-of-the-art results and can substantially improve the delineation of contours near the apex of the heart.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge