Moritz Fuchs
GAUDA: Generative Adaptive Uncertainty-guided Diffusion-based Augmentation for Surgical Segmentation
Jan 18, 2025



Abstract:Augmentation by generative modelling yields a promising alternative to the accumulation of surgical data, where ethical, organisational and regulatory aspects must be considered. Yet, the joint synthesis of (image, mask) pairs for segmentation, a major application in surgery, is rather unexplored. We propose to learn semantically comprehensive yet compact latent representations of the (image, mask) space, which we jointly model with a Latent Diffusion Model. We show that our approach can effectively synthesise unseen high-quality paired segmentation data of remarkable semantic coherence. Generative augmentation is typically applied pre-training by synthesising a fixed number of additional training samples to improve downstream task models. To enhance this approach, we further propose Generative Adaptive Uncertainty-guided Diffusion-based Augmentation (GAUDA), leveraging the epistemic uncertainty of a Bayesian downstream model for targeted online synthesis. We condition the generative model on classes with high estimated uncertainty during training to produce additional unseen samples for these classes. By adaptively utilising the generative model online, we can minimise the number of additional training samples and centre them around the currently most uncertain parts of the data distribution. GAUDA effectively improves downstream segmentation results over comparable methods by an average absolute IoU of 1.6% on CaDISv2 and 1.5% on CholecSeg8k, two prominent surgical datasets for semantic segmentation.
Federated-Continual Dynamic Segmentation of Histopathology guided by Barlow Continuity
Jan 08, 2025



Abstract:Federated- and Continual Learning have been established as approaches to enable privacy-aware learning on continuously changing data, as required for deploying AI systems in histopathology images. However, data shifts can occur in a dynamic world, spatially between institutions and temporally, due to changing data over time. This leads to two issues: Client Drift, where the central model degrades from aggregating data from clients trained on shifted data, and Catastrophic Forgetting, from temporal shifts such as changes in patient populations. Both tend to degrade the model's performance of previously seen data or spatially distributed training. Despite both problems arising from the same underlying problem of data shifts, existing research addresses them only individually. In this work, we introduce a method that can jointly alleviate Client Drift and Catastrophic Forgetting by using our proposed Dynamic Barlow Continuity that evaluates client updates on a public reference dataset and uses this to guide the training process to a spatially and temporally shift-invariant model. We evaluate our approach on the histopathology datasets BCSS and Semicol and prove our method to be highly effective by jointly improving the dice score as much as from 15.8% to 71.6% in Client Drift and from 42.5% to 62.8% in Catastrophic Forgetting. This enables Dynamic Learning by establishing spatio-temporal shift-invariance.
From Pointwise to Powerhouse: Initialising Neural Networks with Generative Models
Oct 25, 2023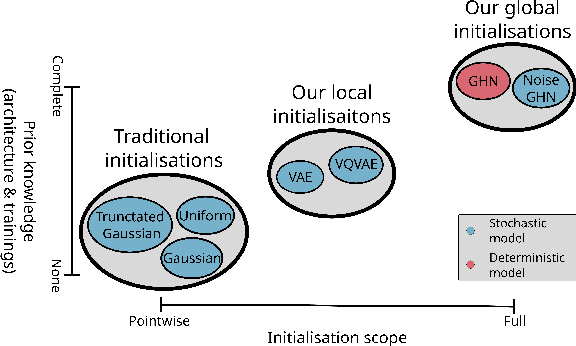
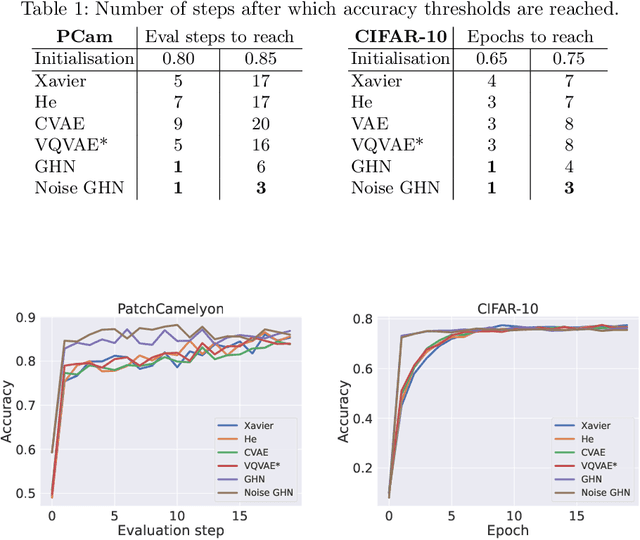
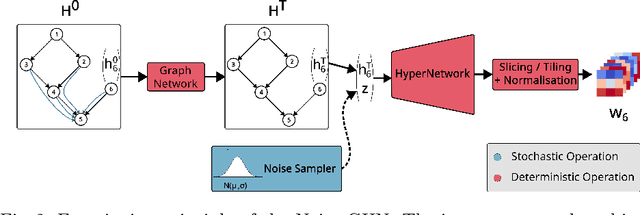
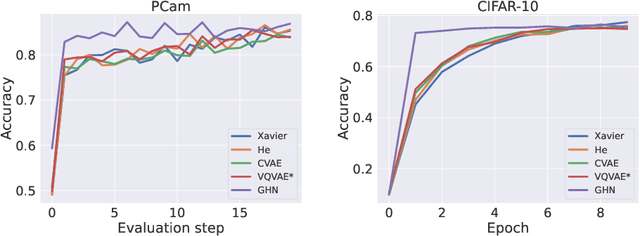
Abstract:Traditional initialisation methods, e.g. He and Xavier, have been effective in avoiding the problem of vanishing or exploding gradients in neural networks. However, they only use simple pointwise distributions, which model one-dimensional variables. Moreover, they ignore most information about the architecture and disregard past training experiences. These limitations can be overcome by employing generative models for initialisation. In this paper, we introduce two groups of new initialisation methods. First, we locally initialise weight groups by employing variational autoencoders. Secondly, we globally initialise full weight sets by employing graph hypernetworks. We thoroughly evaluate the impact of the employed generative models on state-of-the-art neural networks in terms of accuracy, convergence speed and ensembling. Our results show that global initialisations result in higher accuracy and faster initial convergence speed. However, the implementation through graph hypernetworks leads to diminished ensemble performance on out of distribution data. To counteract, we propose a modification called noise graph hypernetwork, which encourages diversity in the produced ensemble members. Furthermore, our approach might be able to transfer learned knowledge to different image distributions. Our work provides insights into the potential, the trade-offs and possible modifications of these new initialisation methods.
Exploring SAM Ablations for Enhancing Medical Segmentation in Radiology and Pathology
Sep 30, 2023



Abstract:Medical imaging plays a critical role in the diagnosis and treatment planning of various medical conditions, with radiology and pathology heavily reliant on precise image segmentation. The Segment Anything Model (SAM) has emerged as a promising framework for addressing segmentation challenges across different domains. In this white paper, we delve into SAM, breaking down its fundamental components and uncovering the intricate interactions between them. We also explore the fine-tuning of SAM and assess its profound impact on the accuracy and reliability of segmentation results, focusing on applications in radiology (specifically, brain tumor segmentation) and pathology (specifically, breast cancer segmentation). Through a series of carefully designed experiments, we analyze SAM's potential application in the field of medical imaging. We aim to bridge the gap between advanced segmentation techniques and the demanding requirements of healthcare, shedding light on SAM's transformative capabilities.
Jointly Exploring Client Drift and Catastrophic Forgetting in Dynamic Learning
Sep 01, 2023



Abstract:Federated and Continual Learning have emerged as potential paradigms for the robust and privacy-aware use of Deep Learning in dynamic environments. However, Client Drift and Catastrophic Forgetting are fundamental obstacles to guaranteeing consistent performance. Existing work only addresses these problems separately, which neglects the fact that the root cause behind both forms of performance deterioration is connected. We propose a unified analysis framework for building a controlled test environment for Client Drift -- by perturbing a defined ratio of clients -- and Catastrophic Forgetting -- by shifting all clients with a particular strength. Our framework further leverages this new combined analysis by generating a 3D landscape of the combined performance impact from both. We demonstrate that the performance drop through Client Drift, caused by a certain share of shifted clients, is correlated to the drop from Catastrophic Forgetting resulting from a corresponding shift strength. Correlation tests between both problems for Computer Vision (CelebA) and Medical Imaging (PESO) support this new perspective, with an average Pearson rank correlation coefficient of over 0.94. Our framework's novel ability of combined spatio-temporal shift analysis allows us to investigate how both forms of distribution shift behave in mixed scenarios, opening a new pathway for better generalization. We show that a combination of moderate Client Drift and Catastrophic Forgetting can even improve the performance of the resulting model (causing a "Generalization Bump") compared to when only one of the shifts occurs individually. We apply a simple and commonly used method from Continual Learning in the federated setting and observe this phenomenon to be reoccurring, leveraging the ability of our framework to analyze existing and novel methods for Federated and Continual Learning.
Synthesising Rare Cataract Surgery Samples with Guided Diffusion Models
Aug 03, 2023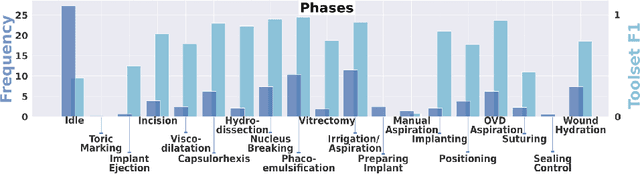
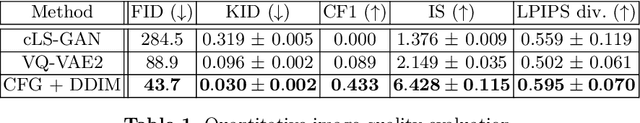
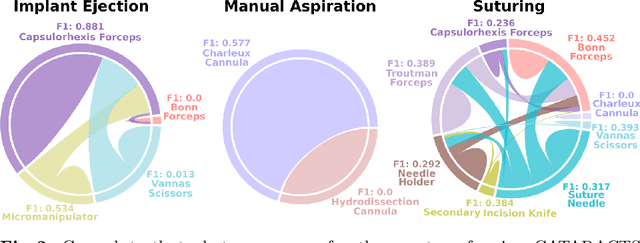
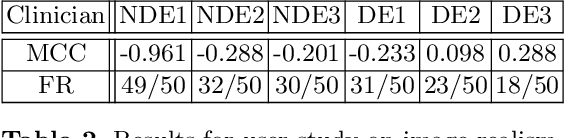
Abstract:Cataract surgery is a frequently performed procedure that demands automation and advanced assistance systems. However, gathering and annotating data for training such systems is resource intensive. The publicly available data also comprises severe imbalances inherent to the surgical process. Motivated by this, we analyse cataract surgery video data for the worst-performing phases of a pre-trained downstream tool classifier. The analysis demonstrates that imbalances deteriorate the classifier's performance on underrepresented cases. To address this challenge, we utilise a conditional generative model based on Denoising Diffusion Implicit Models (DDIM) and Classifier-Free Guidance (CFG). Our model can synthesise diverse, high-quality examples based on complex multi-class multi-label conditions, such as surgical phases and combinations of surgical tools. We affirm that the synthesised samples display tools that the classifier recognises. These samples are hard to differentiate from real images, even for clinical experts with more than five years of experience. Further, our synthetically extended data can improve the data sparsity problem for the downstream task of tool classification. The evaluations demonstrate that the model can generate valuable unseen examples, allowing the tool classifier to improve by up to 10% for rare cases. Overall, our approach can facilitate the development of automated assistance systems for cataract surgery by providing a reliable source of realistic synthetic data, which we make available for everyone.
Federated Stain Normalization for Computational Pathology
Sep 29, 2022Abstract:Although deep federated learning has received much attention in recent years, progress has been made mainly in the context of natural images and barely for computational pathology. However, deep federated learning is an opportunity to create datasets that reflect the data diversity of many laboratories. Further, the effort of dataset construction can be divided among many. Unfortunately, existing algorithms cannot be easily applied to computational pathology since previous work presupposes that data distributions of laboratories must be similar. This is an unlikely assumption, mainly since different laboratories have different staining styles. As a solution, we propose BottleGAN, a generative model that can computationally align the staining styles of many laboratories and can be trained in a privacy-preserving manner to foster federated learning in computational pathology. We construct a heterogenic multi-institutional dataset based on the PESO segmentation dataset and improve the IOU by 42\% compared to existing federated learning algorithms. An implementation of BottleGAN is available at https://github.com/MECLabTUDA/BottleGAN
Distance-based detection of out-of-distribution silent failures for Covid-19 lung lesion segmentation
Aug 05, 2022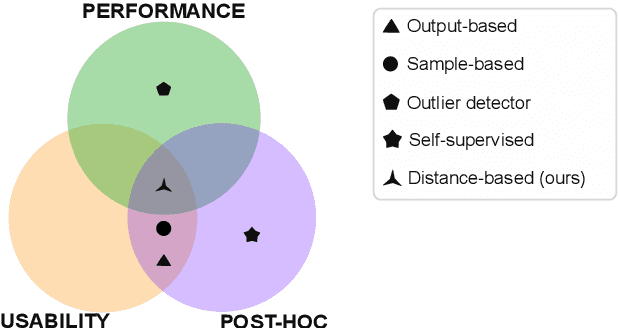
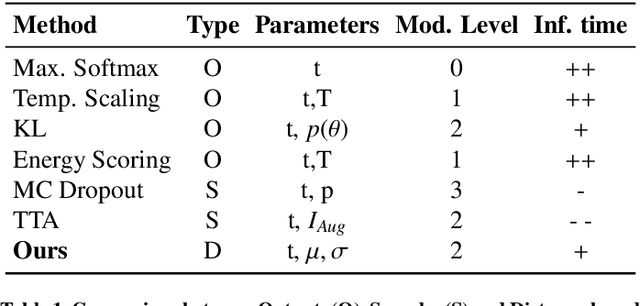
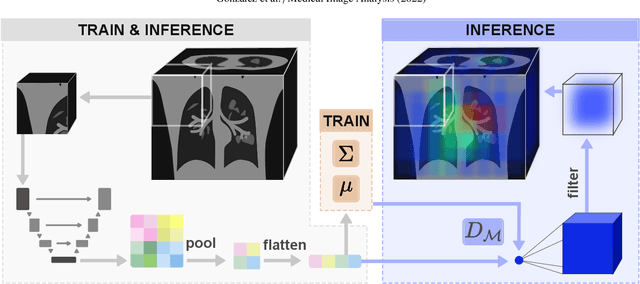
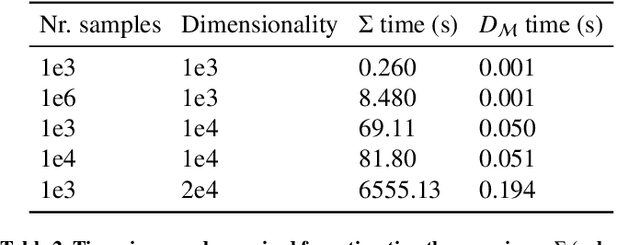
Abstract:Automatic segmentation of ground glass opacities and consolidations in chest computer tomography (CT) scans can potentially ease the burden of radiologists during times of high resource utilisation. However, deep learning models are not trusted in the clinical routine due to failing silently on out-of-distribution (OOD) data. We propose a lightweight OOD detection method that leverages the Mahalanobis distance in the feature space and seamlessly integrates into state-of-the-art segmentation pipelines. The simple approach can even augment pre-trained models with clinically relevant uncertainty quantification. We validate our method across four chest CT distribution shifts and two magnetic resonance imaging applications, namely segmentation of the hippocampus and the prostate. Our results show that the proposed method effectively detects far- and near-OOD samples across all explored scenarios.
FrOoDo: Framework for Out-of-Distribution Detection
Aug 01, 2022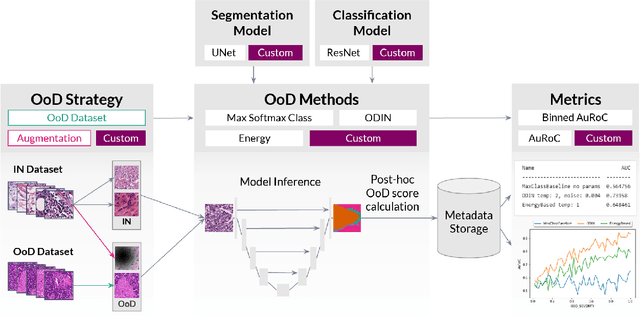
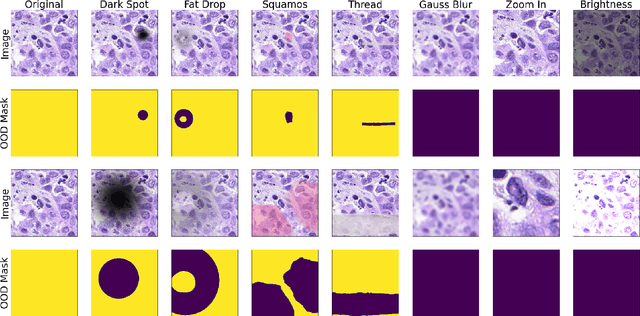
Abstract:FrOoDo is an easy-to-use and flexible framework for Out-of-Distribution detection tasks in digital pathology. It can be used with PyTorch classification and segmentation models, and its modular design allows for easy extension. The goal is to automate the task of OoD Evaluation such that research can focus on the main goal of either designing new models, new methods or evaluating a new dataset. The code can be found at https://github.com/MECLabTUDA/FrOoDo.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge