Hendrik A. Mehrtens
Pathologist-like explainable AI for interpretable Gleason grading in prostate cancer
Oct 19, 2024



Abstract:The aggressiveness of prostate cancer, the most common cancer in men worldwide, is primarily assessed based on histopathological data using the Gleason scoring system. While artificial intelligence (AI) has shown promise in accurately predicting Gleason scores, these predictions often lack inherent explainability, potentially leading to distrust in human-machine interactions. To address this issue, we introduce a novel dataset of 1,015 tissue microarray core images, annotated by an international group of 54 pathologists. The annotations provide detailed localized pattern descriptions for Gleason grading in line with international guidelines. Utilizing this dataset, we develop an inherently explainable AI system based on a U-Net architecture that provides predictions leveraging pathologists' terminology. This approach circumvents post-hoc explainability methods while maintaining or exceeding the performance of methods trained directly for Gleason pattern segmentation (Dice score: 0.713 $\pm$ 0.003 trained on explanations vs. 0.691 $\pm$ 0.010 trained on Gleason patterns). By employing soft labels during training, we capture the intrinsic uncertainty in the data, yielding strong results in Gleason pattern segmentation even in the context of high interobserver variability. With the release of this dataset, we aim to encourage further research into segmentation in medical tasks with high levels of subjectivity and to advance the understanding of pathologists' reasoning processes.
On the calibration of neural networks for histological slide-level classification
Dec 15, 2023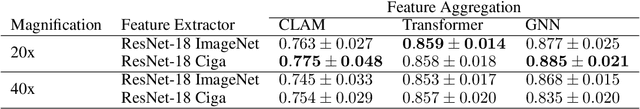
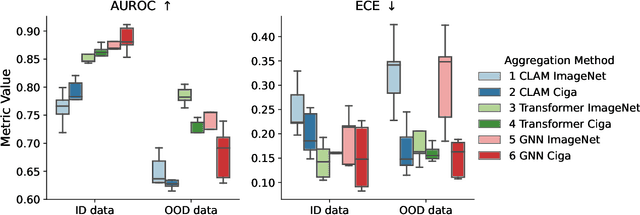
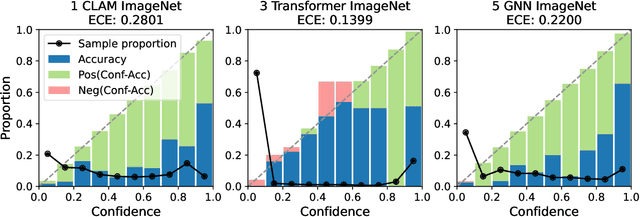
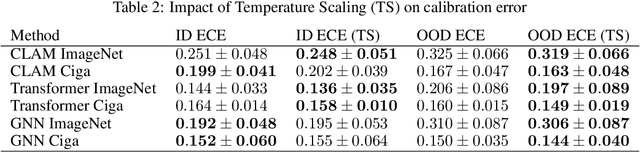
Abstract:Deep Neural Networks have shown promising classification performance when predicting certain biomarkers from Whole Slide Images in digital pathology. However, the calibration of the networks' output probabilities is often not evaluated. Communicating uncertainty by providing reliable confidence scores is of high relevance in the medical context. In this work, we compare three neural network architectures that combine feature representations on patch-level to a slide-level prediction with respect to their classification performance and evaluate their calibration. As slide-level classification task, we choose the prediction of Microsatellite Instability from Colorectal Cancer tissue sections. We observe that Transformers lead to good results in terms of classification performance and calibration. When evaluating the classification performance on a separate dataset, we observe that Transformers generalize best. The investigation of reliability diagrams provides additional insights to the Expected Calibration Error metric and we observe that especially Transformers push the output probabilities to extreme values, which results in overconfident predictions.
Benchmarking common uncertainty estimation methods with histopathological images under domain shift and label noise
Jan 03, 2023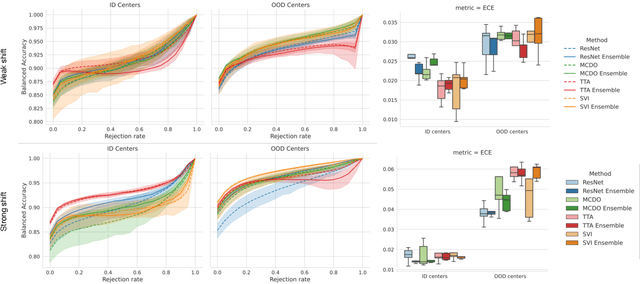
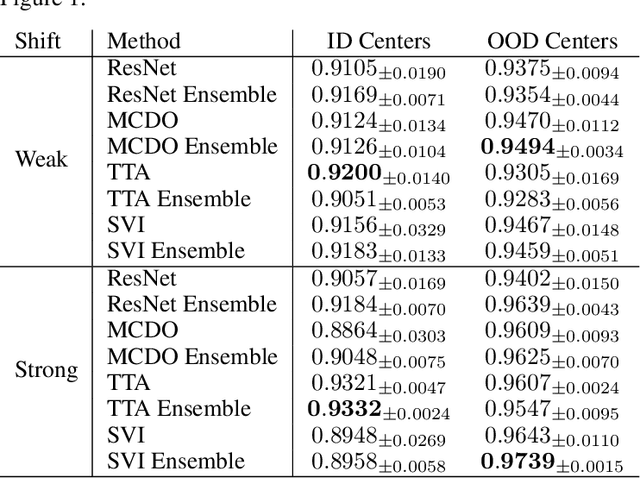
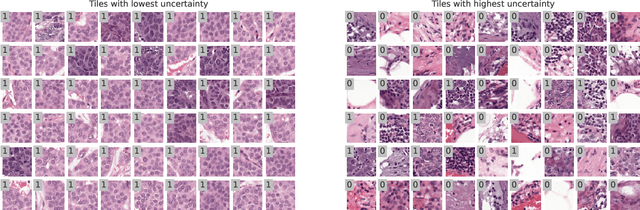
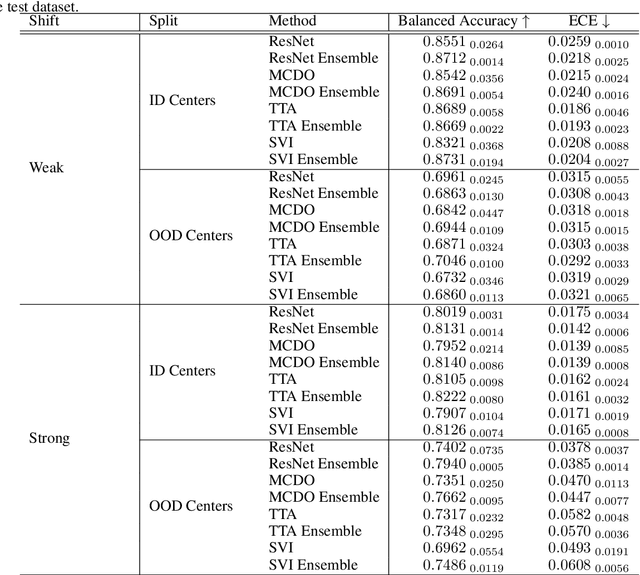
Abstract:In the past years, deep learning has seen an increase of usage in the domain of histopathological applications. However, while these approaches have shown great potential, in high-risk environments deep learning models need to be able to judge their own uncertainty and be able to reject inputs when there is a significant chance of misclassification. In this work, we conduct a rigorous evaluation of the most commonly used uncertainty and robustness methods for the classification of Whole-Slide-Images under domain shift using the H\&E stained Camelyon17 breast cancer dataset. Although it is known that histopathological data can be subject to strong domain shift and label noise, to our knowledge this is the first work that compares the most common methods for uncertainty estimation under these aspects. In our experiments, we compare Stochastic Variational Inference, Monte-Carlo Dropout, Deep Ensembles, Test-Time Data Augmentation as well as combinations thereof. We observe that ensembles of methods generally lead to higher accuracies and better calibration and that Test-Time Data Augmentation can be a promising alternative when choosing an appropriate set of augmentations. Across methods, a rejection of the most uncertain tiles leads to a significant increase in classification accuracy on both in-distribution as well as out-of-distribution data. Furthermore, we conduct experiments comparing these methods under varying conditions of label noise. We observe that the border regions of the Camelyon17 dataset are subject to label noise and evaluate the robustness of the included methods against different noise levels. Lastly, we publish our code framework to facilitate further research on uncertainty estimation on histopathological data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge