Jens Rittscher
Histology-informed tiling of whole tissue sections improves the interpretability and predictability of cancer relapse and genetic alterations
Nov 13, 2025
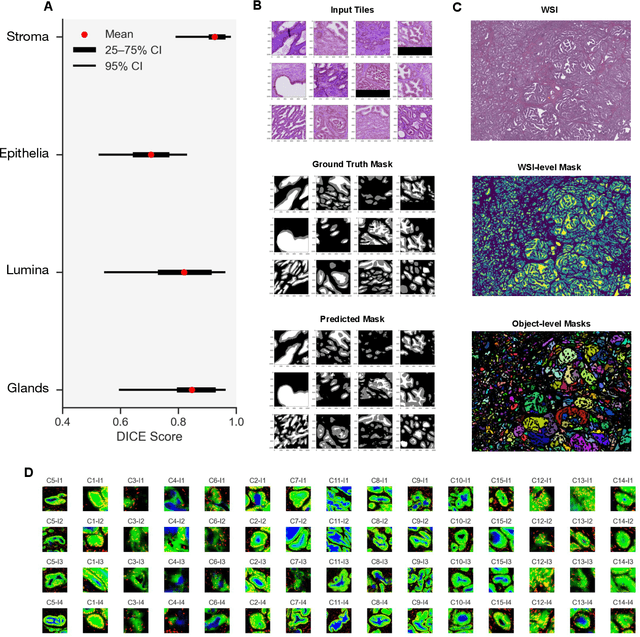
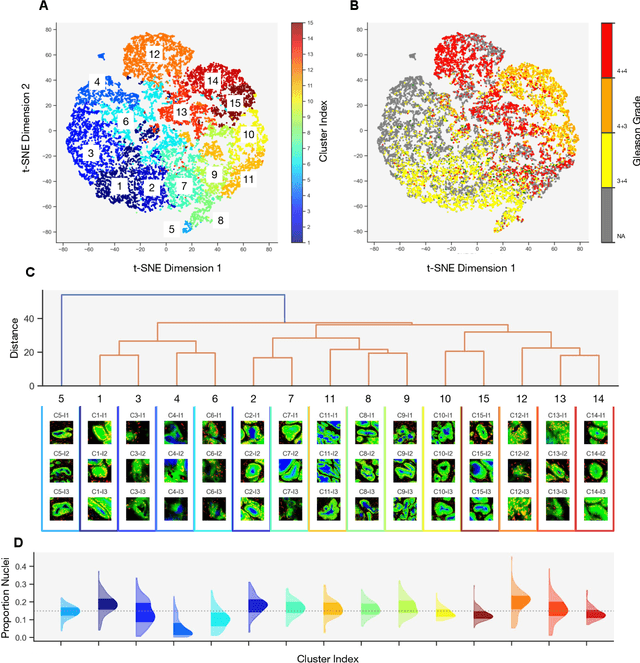
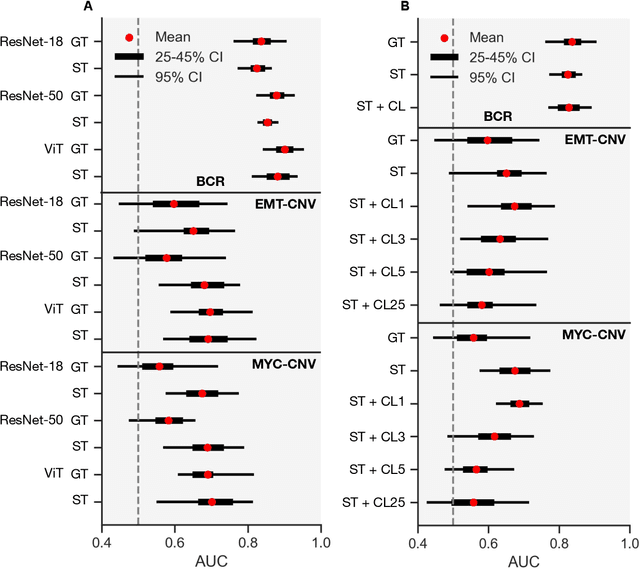
Abstract:Histopathologists establish cancer grade by assessing histological structures, such as glands in prostate cancer. Yet, digital pathology pipelines often rely on grid-based tiling that ignores tissue architecture. This introduces irrelevant information and limits interpretability. We introduce histology-informed tiling (HIT), which uses semantic segmentation to extract glands from whole slide images (WSIs) as biologically meaningful input patches for multiple-instance learning (MIL) and phenotyping. Trained on 137 samples from the ProMPT cohort, HIT achieved a gland-level Dice score of 0.83 +/- 0.17. By extracting 380,000 glands from 760 WSIs across ICGC-C and TCGA-PRAD cohorts, HIT improved MIL models AUCs by 10% for detecting copy number variation (CNVs) in genes related to epithelial-mesenchymal transitions (EMT) and MYC, and revealed 15 gland clusters, several of which were associated with cancer relapse, oncogenic mutations, and high Gleason. Therefore, HIT improved the accuracy and interpretability of MIL predictions, while streamlining computations by focussing on biologically meaningful structures during feature extraction.
AdaFusion: Prompt-Guided Inference with Adaptive Fusion of Pathology Foundation Models
Aug 07, 2025Abstract:Pathology foundation models (PFMs) have demonstrated strong representational capabilities through self-supervised pre-training on large-scale, unannotated histopathology image datasets. However, their diverse yet opaque pretraining contexts, shaped by both data-related and structural/training factors, introduce latent biases that hinder generalisability and transparency in downstream applications. In this paper, we propose AdaFusion, a novel prompt-guided inference framework that, to our knowledge, is among the very first to dynamically integrate complementary knowledge from multiple PFMs. Our method compresses and aligns tile-level features from diverse models and employs a lightweight attention mechanism to adaptively fuse them based on tissue phenotype context. We evaluate AdaFusion on three real-world benchmarks spanning treatment response prediction, tumour grading, and spatial gene expression inference. Our approach consistently surpasses individual PFMs across both classification and regression tasks, while offering interpretable insights into each model's biosemantic specialisation. These results highlight AdaFusion's ability to bridge heterogeneous PFMs, achieving both enhanced performance and interpretability of model-specific inductive biases.
Self-supervised Monocular Depth and Pose Estimation for Endoscopy with Generative Latent Priors
Nov 26, 2024


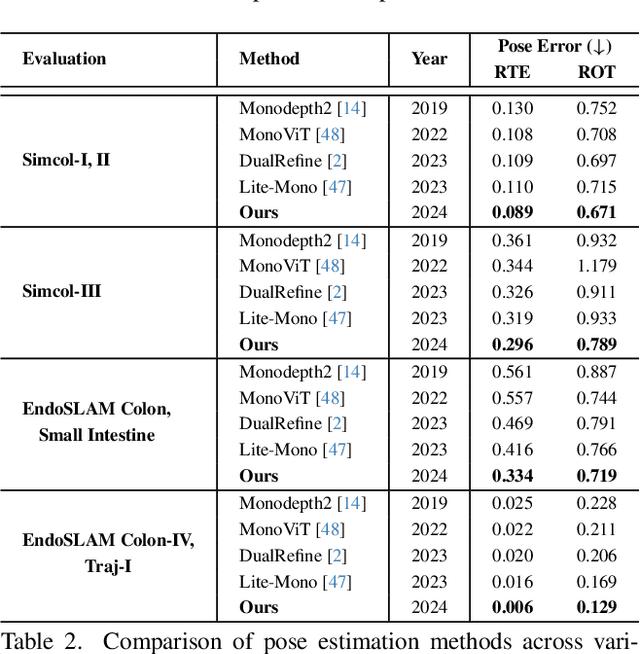
Abstract:Accurate 3D mapping in endoscopy enables quantitative, holistic lesion characterization within the gastrointestinal (GI) tract, requiring reliable depth and pose estimation. However, endoscopy systems are monocular, and existing methods relying on synthetic datasets or complex models often lack generalizability in challenging endoscopic conditions. We propose a robust self-supervised monocular depth and pose estimation framework that incorporates a Generative Latent Bank and a Variational Autoencoder (VAE). The Generative Latent Bank leverages extensive depth scenes from natural images to condition the depth network, enhancing realism and robustness of depth predictions through latent feature priors. For pose estimation, we reformulate it within a VAE framework, treating pose transitions as latent variables to regularize scale, stabilize z-axis prominence, and improve x-y sensitivity. This dual refinement pipeline enables accurate depth and pose predictions, effectively addressing the GI tract's complex textures and lighting. Extensive evaluations on SimCol and EndoSLAM datasets confirm our framework's superior performance over published self-supervised methods in endoscopic depth and pose estimation.
SSTFB: Leveraging self-supervised pretext learning and temporal self-attention with feature branching for real-time video polyp segmentation
Jun 14, 2024Abstract:Polyps are early cancer indicators, so assessing occurrences of polyps and their removal is critical. They are observed through a colonoscopy screening procedure that generates a stream of video frames. Segmenting polyps in their natural video screening procedure has several challenges, such as the co-existence of imaging artefacts, motion blur, and floating debris. Most existing polyp segmentation algorithms are developed on curated still image datasets that do not represent real-world colonoscopy. Their performance often degrades on video data. We propose a video polyp segmentation method that performs self-supervised learning as an auxiliary task and a spatial-temporal self-attention mechanism for improved representation learning. Our end-to-end configuration and joint optimisation of losses enable the network to learn more discriminative contextual features in videos. Our experimental results demonstrate an improvement with respect to several state-of-the-art (SOTA) methods. Our ablation study also confirms that the choice of the proposed joint end-to-end training improves network accuracy by over 3% and nearly 10% on both the Dice similarity coefficient and intersection-over-union compared to the recently proposed method PNS+ and Polyp-PVT, respectively. Results on previously unseen video data indicate that the proposed method generalises.
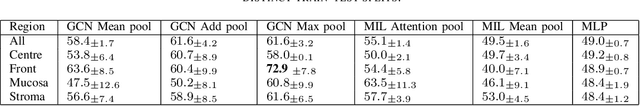
Beyond attention: deriving biologically interpretable insights from weakly-supervised multiple-instance learning models
Sep 07, 2023



Abstract:Recent advances in attention-based multiple instance learning (MIL) have improved our insights into the tissue regions that models rely on to make predictions in digital pathology. However, the interpretability of these approaches is still limited. In particular, they do not report whether high-attention regions are positively or negatively associated with the class labels or how well these regions correspond to previously established clinical and biological knowledge. We address this by introducing a post-training methodology to analyse MIL models. Firstly, we introduce prediction-attention-weighted (PAW) maps by combining tile-level attention and prediction scores produced by a refined encoder, allowing us to quantify the predictive contribution of high-attention regions. Secondly, we introduce a biological feature instantiation technique by integrating PAW maps with nuclei segmentation masks. This further improves interpretability by providing biologically meaningful features related to the cellular organisation of the tissue and facilitates comparisons with known clinical features. We illustrate the utility of our approach by comparing PAW maps obtained for prostate cancer diagnosis (i.e. samples containing malignant tissue, 381/516 tissue samples) and prognosis (i.e. samples from patients with biochemical recurrence following surgery, 98/663 tissue samples) in a cohort of patients from the international cancer genome consortium (ICGC UK Prostate Group). Our approach reveals that regions that are predictive of adverse prognosis do not tend to co-locate with the tumour regions, indicating that non-cancer cells should also be studied when evaluating prognosis.
SSL-CPCD: Self-supervised learning with composite pretext-class discrimination for improved generalisability in endoscopic image analysis
May 31, 2023Abstract:Data-driven methods have shown tremendous progress in medical image analysis. In this context, deep learning-based supervised methods are widely popular. However, they require a large amount of training data and face issues in generalisability to unseen datasets that hinder clinical translation. Endoscopic imaging data incorporates large inter- and intra-patient variability that makes these models more challenging to learn representative features for downstream tasks. Thus, despite the publicly available datasets and datasets that can be generated within hospitals, most supervised models still underperform. While self-supervised learning has addressed this problem to some extent in natural scene data, there is a considerable performance gap in the medical image domain. In this paper, we propose to explore patch-level instance-group discrimination and penalisation of inter-class variation using additive angular margin within the cosine similarity metrics. Our novel approach enables models to learn to cluster similar representative patches, thereby improving their ability to provide better separation between different classes. Our results demonstrate significant improvement on all metrics over the state-of-the-art (SOTA) methods on the test set from the same and diverse datasets. We evaluated our approach for classification, detection, and segmentation. SSL-CPCD achieves 79.77% on Top 1 accuracy for ulcerative colitis classification, 88.62% on mAP for polyp detection, and 82.32% on dice similarity coefficient for segmentation tasks are nearly over 4%, 2%, and 3%, respectively, compared to the baseline architectures. We also demonstrate that our method generalises better than all SOTA methods to unseen datasets, reporting nearly 7% improvement in our generalisability assessment.
Patch-level instance-group discrimination with pretext-invariant learning for colitis scoring
Jul 11, 2022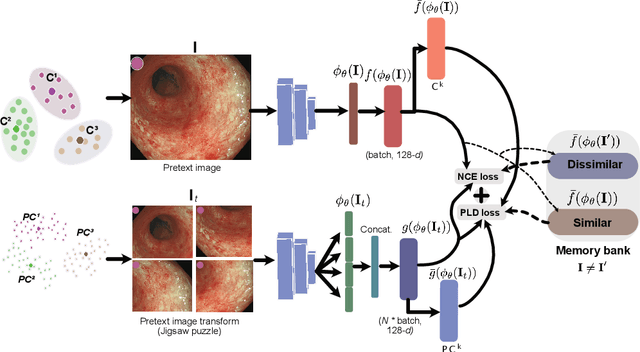
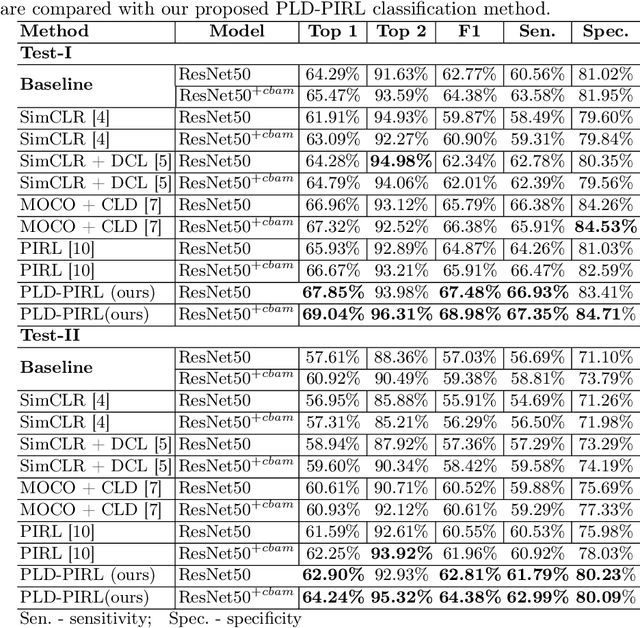
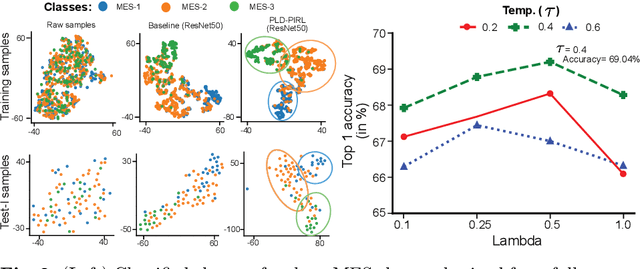
Abstract:Inflammatory bowel disease (IBD), in particular ulcerative colitis (UC), is graded by endoscopists and this assessment is the basis for risk stratification and therapy monitoring. Presently, endoscopic characterisation is largely operator dependant leading to sometimes undesirable clinical outcomes for patients with IBD. We focus on the Mayo Endoscopic Scoring (MES) system which is widely used but requires the reliable identification of subtle changes in mucosal inflammation. Most existing deep learning classification methods cannot detect these fine-grained changes which make UC grading such a challenging task. In this work, we introduce a novel patch-level instance-group discrimination with pretext-invariant representation learning (PLD-PIRL) for self-supervised learning (SSL). Our experiments demonstrate both improved accuracy and robustness compared to the baseline supervised network and several state-of-the-art SSL methods. Compared to the baseline (ResNet50) supervised classification our proposed PLD-PIRL obtained an improvement of 4.75% on hold-out test data and 6.64% on unseen center test data for top-1 accuracy.
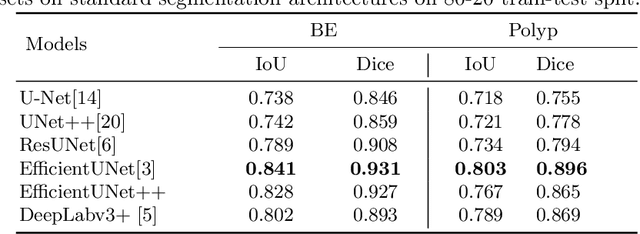
Assessing generalisability of deep learning-based polyp detection and segmentation methods through a computer vision challenge
Feb 24, 2022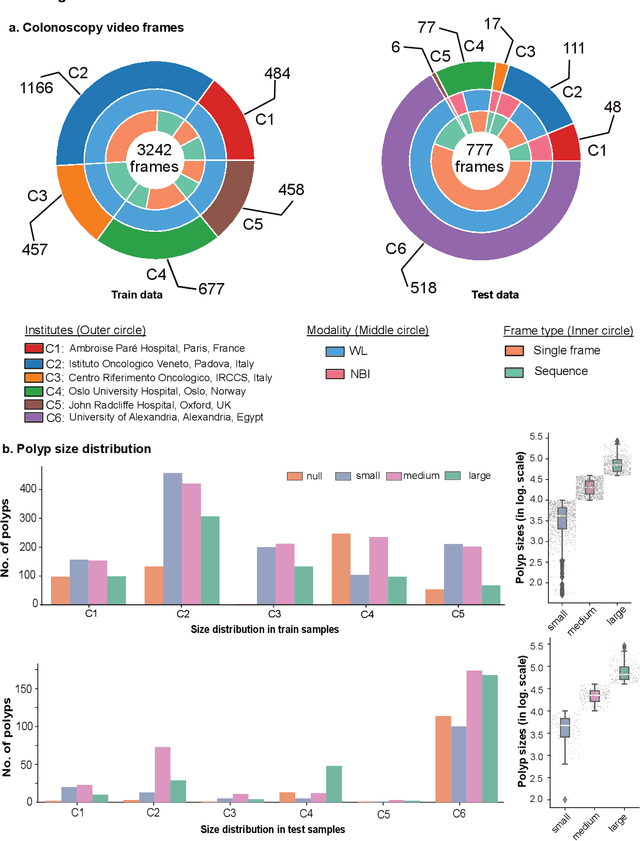
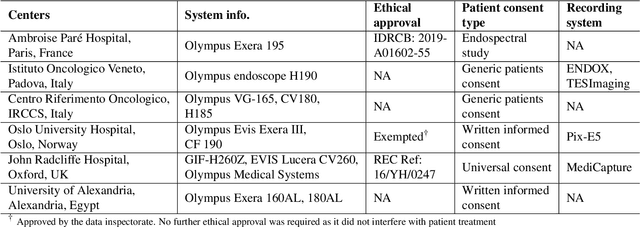
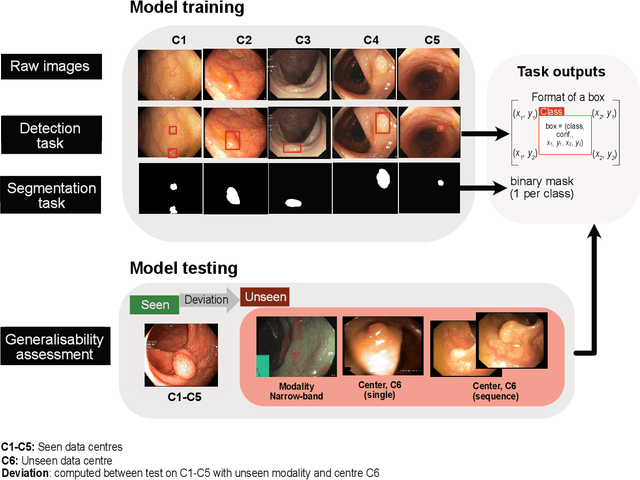
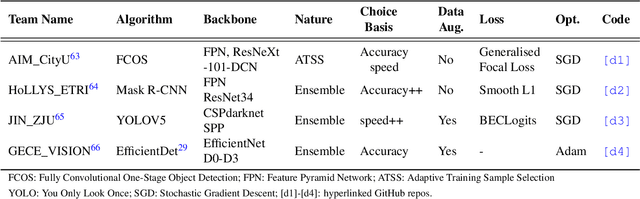
Abstract:Polyps are well-known cancer precursors identified by colonoscopy. However, variability in their size, location, and surface largely affect identification, localisation, and characterisation. Moreover, colonoscopic surveillance and removal of polyps (referred to as polypectomy ) are highly operator-dependent procedures. There exist a high missed detection rate and incomplete removal of colonic polyps due to their variable nature, the difficulties to delineate the abnormality, the high recurrence rates, and the anatomical topography of the colon. There have been several developments in realising automated methods for both detection and segmentation of these polyps using machine learning. However, the major drawback in most of these methods is their ability to generalise to out-of-sample unseen datasets that come from different centres, modalities and acquisition systems. To test this hypothesis rigorously we curated a multi-centre and multi-population dataset acquired from multiple colonoscopy systems and challenged teams comprising machine learning experts to develop robust automated detection and segmentation methods as part of our crowd-sourcing Endoscopic computer vision challenge (EndoCV) 2021. In this paper, we analyse the detection results of the four top (among seven) teams and the segmentation results of the five top teams (among 16). Our analyses demonstrate that the top-ranking teams concentrated on accuracy (i.e., accuracy > 80% on overall Dice score on different validation sets) over real-time performance required for clinical applicability. We further dissect the methods and provide an experiment-based hypothesis that reveals the need for improved generalisability to tackle diversity present in multi-centre datasets.
A Graph Based Neural Network Approach to Immune Profiling of Multiplexed Tissue Samples
Feb 01, 2022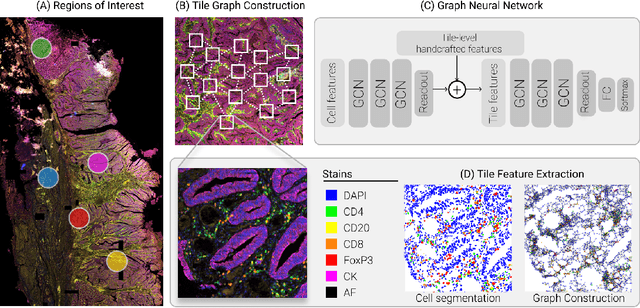
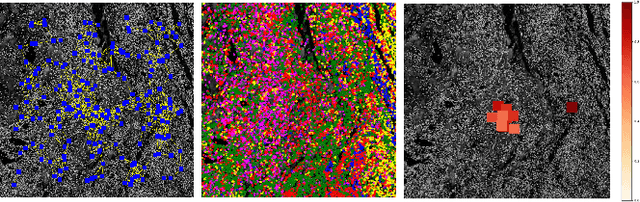
Abstract:Multiplexed immunofluorescence provides an unprecedented opportunity for studying specific cell-to-cell and cell microenvironment interactions. We employ graph neural networks to combine features obtained from tissue morphology with measurements of protein expression to profile the tumour microenvironment associated with different tumour stages. Our framework presents a new approach to analysing and processing these complex multi-dimensional datasets that overcomes some of the key challenges in analysing these data and opens up the opportunity to abstract biologically meaningful interactions.
EndoUDA: A modality independent segmentation approach for endoscopy imaging
Jul 12, 2021
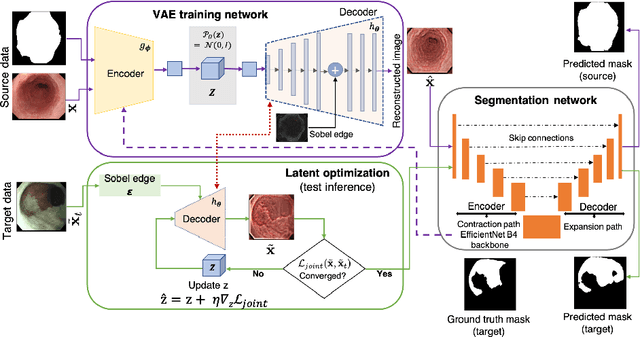
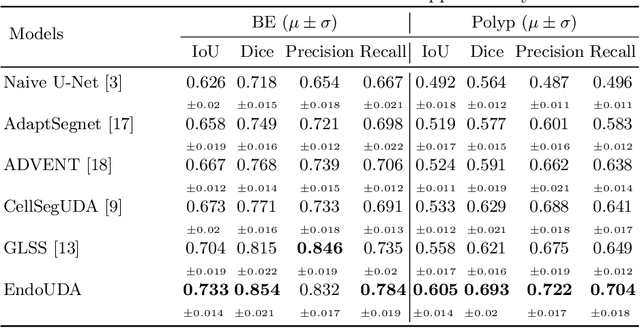
Abstract:Gastrointestinal (GI) cancer precursors require frequent monitoring for risk stratification of patients. Automated segmentation methods can help to assess risk areas more accurately, and assist in therapeutic procedures or even removal. In clinical practice, addition to the conventional white-light imaging (WLI), complimentary modalities such as narrow-band imaging (NBI) and fluorescence imaging are used. While, today most segmentation approaches are supervised and only concentrated on a single modality dataset, this work exploits to use a target-independent unsupervised domain adaptation (UDA) technique that is capable to generalize to an unseen target modality. In this context, we propose a novel UDA-based segmentation method that couples the variational autoencoder and U-Net with a common EfficientNet-B4 backbone, and uses a joint loss for latent-space optimization for target samples. We show that our model can generalize to unseen target NBI (target) modality when trained using only WLI (source) modality. Our experiments on both upper and lower GI endoscopy data show the effectiveness of our approach compared to naive supervised approach and state-of-the-art UDA segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge