Gorkem Polat
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Assessing generalisability of deep learning-based polyp detection and segmentation methods through a computer vision challenge
Feb 24, 2022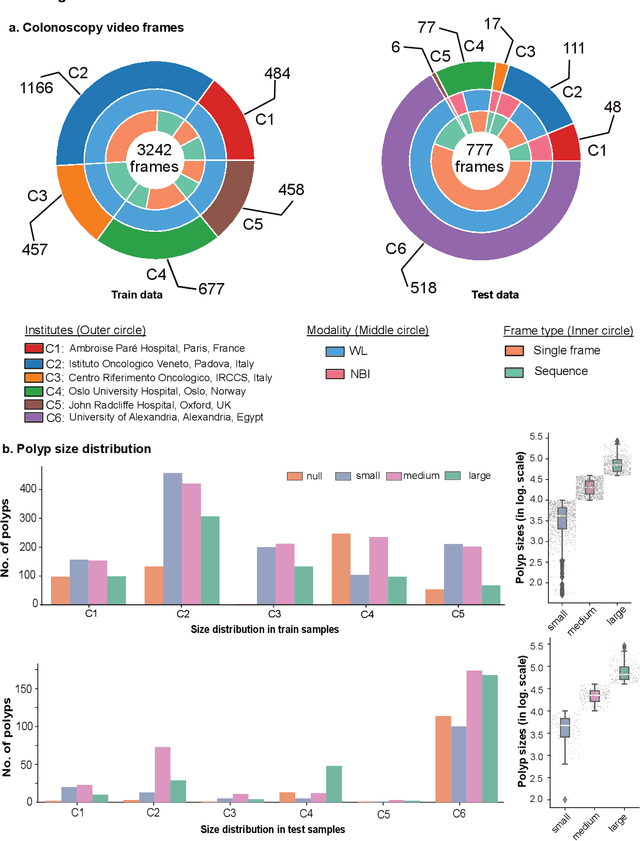
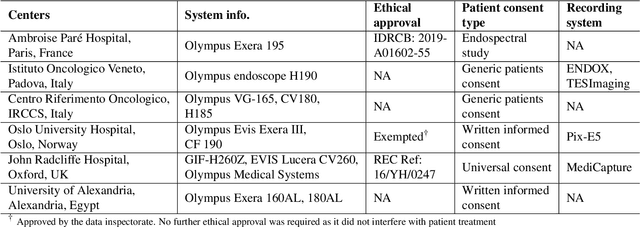
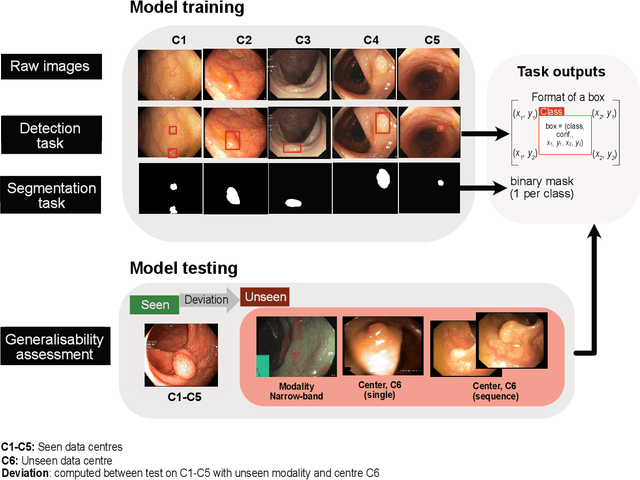
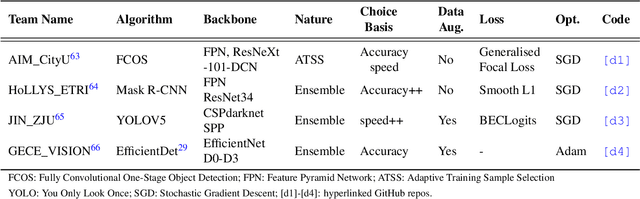
Abstract:Polyps are well-known cancer precursors identified by colonoscopy. However, variability in their size, location, and surface largely affect identification, localisation, and characterisation. Moreover, colonoscopic surveillance and removal of polyps (referred to as polypectomy ) are highly operator-dependent procedures. There exist a high missed detection rate and incomplete removal of colonic polyps due to their variable nature, the difficulties to delineate the abnormality, the high recurrence rates, and the anatomical topography of the colon. There have been several developments in realising automated methods for both detection and segmentation of these polyps using machine learning. However, the major drawback in most of these methods is their ability to generalise to out-of-sample unseen datasets that come from different centres, modalities and acquisition systems. To test this hypothesis rigorously we curated a multi-centre and multi-population dataset acquired from multiple colonoscopy systems and challenged teams comprising machine learning experts to develop robust automated detection and segmentation methods as part of our crowd-sourcing Endoscopic computer vision challenge (EndoCV) 2021. In this paper, we analyse the detection results of the four top (among seven) teams and the segmentation results of the five top teams (among 16). Our analyses demonstrate that the top-ranking teams concentrated on accuracy (i.e., accuracy > 80% on overall Dice score on different validation sets) over real-time performance required for clinical applicability. We further dissect the methods and provide an experiment-based hypothesis that reveals the need for improved generalisability to tackle diversity present in multi-centre datasets.
Evaluation and Analysis of Different Aggregation and Hyperparameter Selection Methods for Federated Brain Tumor Segmentation
Feb 16, 2022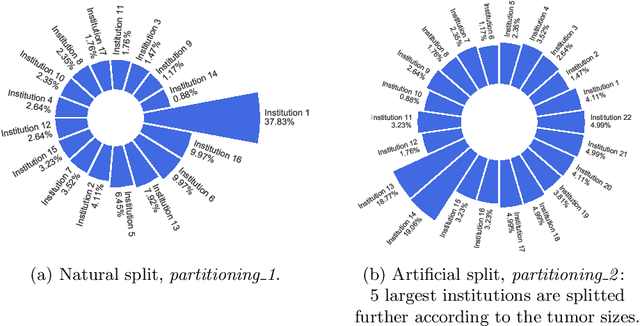
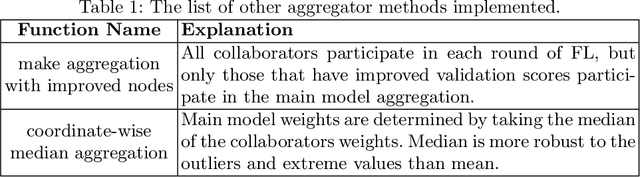

Abstract:Availability of large, diverse, and multi-national datasets is crucial for the development of effective and clinically applicable AI systems in the medical imaging domain. However, forming a global model by bringing these datasets together at a central location, comes along with various data privacy and ownership problems. To alleviate these problems, several recent studies focus on the federated learning paradigm, a distributed learning approach for decentralized data. Federated learning leverages all the available data without any need for sharing collaborators' data with each other or collecting them on a central server. Studies show that federated learning can provide competitive performance with conventional central training, while having a good generalization capability. In this work, we have investigated several federated learning approaches on the brain tumor segmentation problem. We explore different strategies for faster convergence and better performance which can also work on strong Non-IID cases.
Class Distance Weighted Cross-Entropy Loss for Ulcerative Colitis Severity Estimation
Feb 09, 2022

Abstract:Endoscopic Mayo score and Ulcerative Colitis Endoscopic Index of Severity are commonly used scoring systems for the assessment of endoscopic severity of ulcerative colitis. They are based on assigning a score in relation to the disease activity, which creates a rank among the levels, making it an ordinal regression problem. On the other hand, most studies use categorical cross-entropy loss function, which is not optimal for the ordinal regression problem, to train the deep learning models. In this study, we propose a novel loss function called class distance weighted cross-entropy (CDW-CE) that respects the order of the classes and takes the distance of the classes into account in calculation of cost. Experimental evaluations show that CDW-CE outperforms the conventional categorical cross-entropy and CORN framework, which is designed for the ordinal regression problems. In addition, CDW-CE does not require any modifications at the output layer and is compatible with the class activation map visualization techniques.
BARFED: Byzantine Attack-Resistant Federated Averaging Based on Outlier Elimination
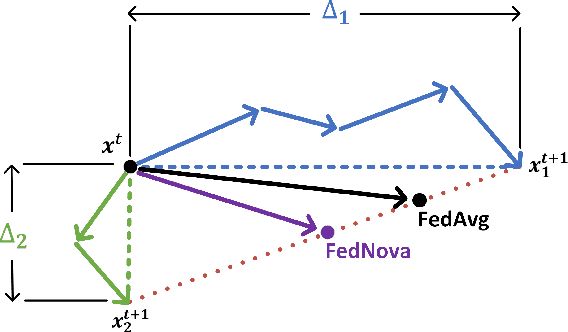
Nov 08, 2021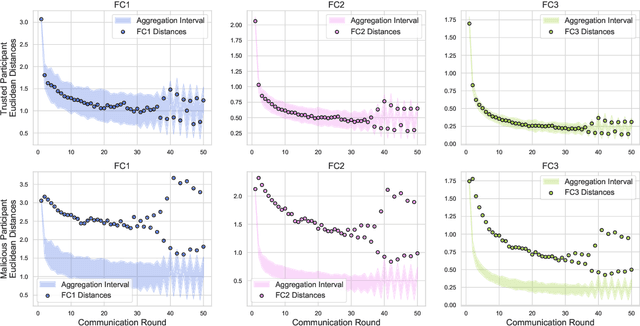
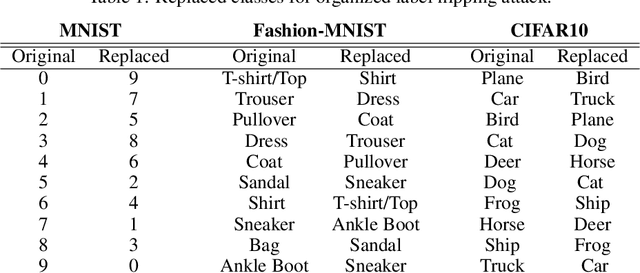
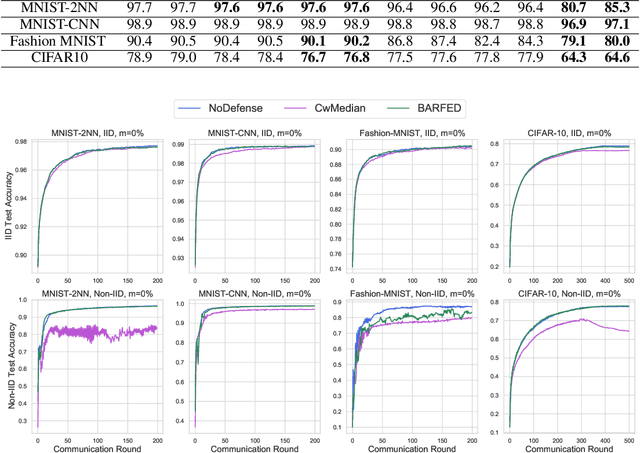
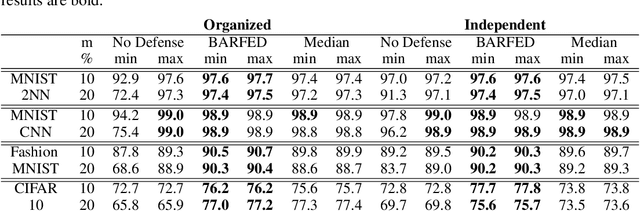
Abstract:In federated learning, each participant trains its local model with its own data and a global model is formed at a trusted server by aggregating model updates coming from these participants. Since the server has no effect and visibility on the training procedure of the participants to ensure privacy, the global model becomes vulnerable to attacks such as data poisoning and model poisoning. Although many defense algorithms have recently been proposed to address these attacks, they often make strong assumptions that do not agree with the nature of federated learning, such as Non-IID datasets. Moreover, they mostly lack comprehensive experimental analyses. In this work, we propose a defense algorithm called BARFED that does not make any assumptions about data distribution, update similarity of participants, or the ratio of the malicious participants. BARFED mainly considers the outlier status of participant updates for each layer of the model architecture based on the distance to the global model. Hence, the participants that do not have any outlier layer are involved in model aggregation. We perform extensive experiments on many grounds and show that the proposed approach provides a robust defense against different attacks.
Effect of Input Size on the Classification of Lung Nodules Using Convolutional Neural Networks
Jul 11, 2021Abstract:Recent studies have shown that lung cancer screening using annual low-dose computed tomography (CT) reduces lung cancer mortality by 20% compared to traditional chest radiography. Therefore, CT lung screening has started to be used widely all across the world. However, analyzing these images is a serious burden for radiologists. The number of slices in a CT scan can be up to 600. Therefore, computer-aided-detection (CAD) systems are very important for faster and more accurate assessment of the data. In this study, we proposed a framework that analyzes CT lung screenings using convolutional neural networks (CNNs) to reduce false positives. We trained our model with different volume sizes and showed that volume size plays a critical role in the performance of the system. We also used different fusions in order to show their power and effect on the overall accuracy. 3D CNNs were preferred over 2D CNNs because 2D convolutional operations applied to 3D data could result in information loss. The proposed framework has been tested on the dataset provided by the LUNA16 Challenge and resulted in a sensitivity of 0.831 at 1 false positive per scan.
A translational pathway of deep learning methods in GastroIntestinal Endoscopy
Oct 12, 2020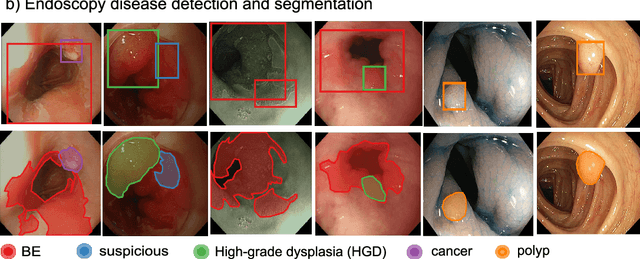
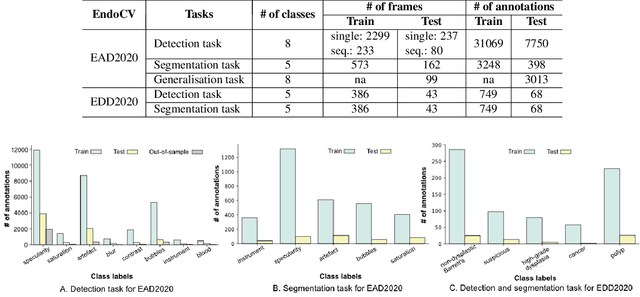
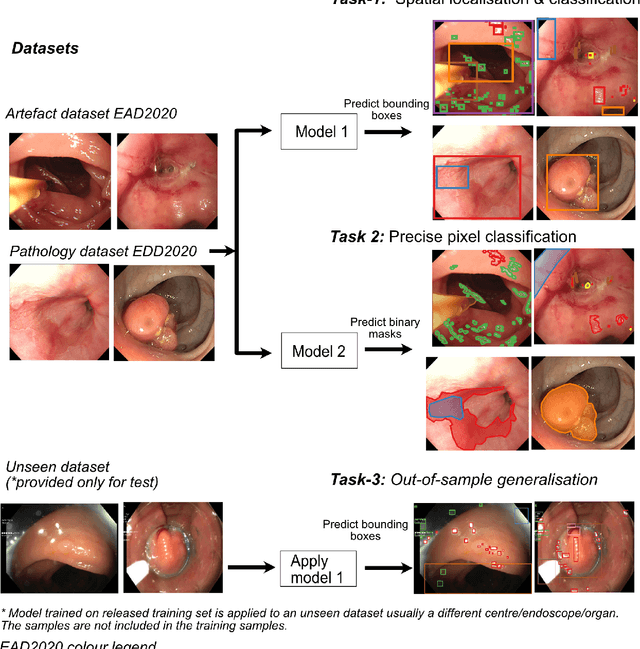
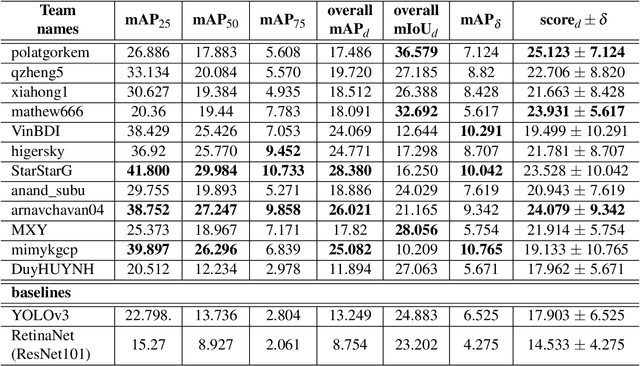
Abstract:The Endoscopy Computer Vision Challenge (EndoCV) is a crowd-sourcing initiative to address eminent problems in developing reliable computer aided detection and diagnosis endoscopy systems and suggest a pathway for clinical translation of technologies. Whilst endoscopy is a widely used diagnostic and treatment tool for hollow-organs, there are several core challenges often faced by endoscopists, mainly: 1) presence of multi-class artefacts that hinder their visual interpretation, and 2) difficulty in identifying subtle precancerous precursors and cancer abnormalities. Artefacts often affect the robustness of deep learning methods applied to the gastrointestinal tract organs as they can be confused with tissue of interest. EndoCV2020 challenges are designed to address research questions in these remits. In this paper, we present a summary of methods developed by the top 17 teams and provide an objective comparison of state-of-the-art methods and methods designed by the participants for two sub-challenges: i) artefact detection and segmentation (EAD2020), and ii) disease detection and segmentation (EDD2020). Multi-center, multi-organ, multi-class, and multi-modal clinical endoscopy datasets were compiled for both EAD2020 and EDD2020 sub-challenges. An out-of-sample generalisation ability of detection algorithms was also evaluated. Whilst most teams focused on accuracy improvements, only a few methods hold credibility for clinical usability. The best performing teams provided solutions to tackle class imbalance, and variabilities in size, origin, modality and occurrences by exploring data augmentation, data fusion, and optimal class thresholding techniques.
False Positive Reduction in Lung Computed Tomography Images using Convolutional Neural Networks
Nov 04, 2018
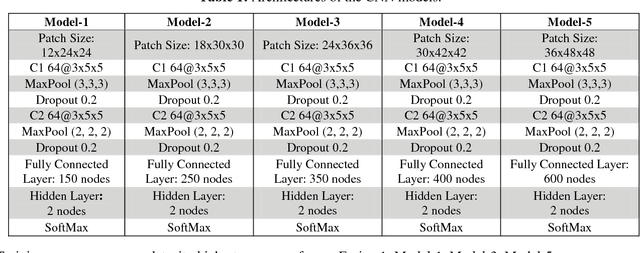
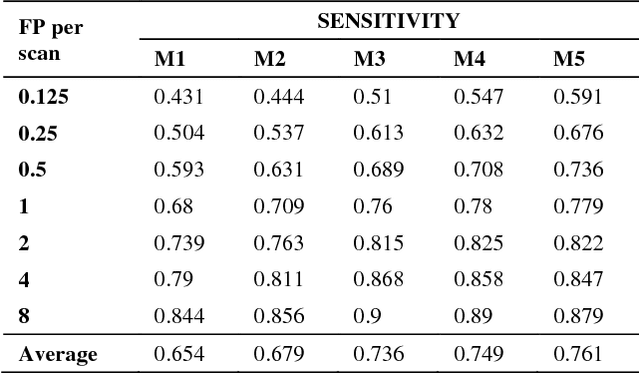
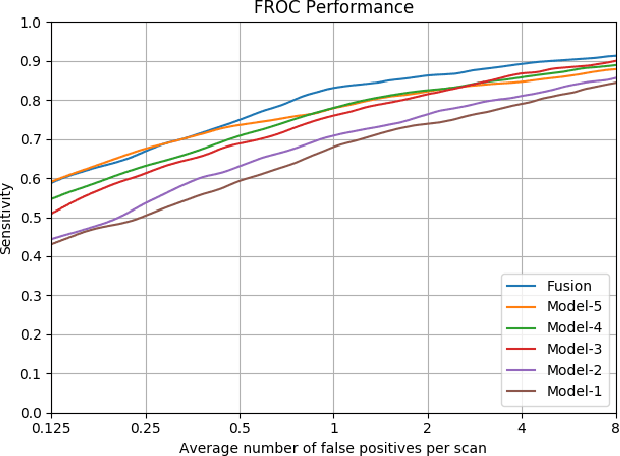
Abstract:Recent studies have shown that lung cancer screening using annual low-dose computed tomography (CT) reduces lung cancer mortality by 20% compared to traditional chest radiography. Therefore, CT lung screening has started to be used widely all across the world. However, analyzing these images is a serious burden for radiologists. In this study, we propose a novel and simple framework that analyzes CT lung screenings using convolutional neural networks (CNNs) and reduces false positives. Our framework shows that even non-complex architectures are very powerful to classify 3D nodule data when compared to traditional methods. We also use different fusions in order to show their power and effect on the overall score. 3D CNNs are preferred over 2D CNNs because data are in 3D, and 2D convolutional operations may result in information loss. Mini-batch is used in order to overcome class-imbalance. Proposed framework has been validated according to the LUNA16 challenge evaluation and got score of 0.786, which is the average sensitivity values at seven predefined false positive (FP) points.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge