Amar Hekalo
QBI: Quantile-based Bias Initialization for Efficient Private Data Reconstruction in Federated Learning
Jun 26, 2024



Abstract:Federated learning enables the training of machine learning models on distributed data without compromising user privacy, as data remains on personal devices and only model updates, such as gradients, are shared with a central coordinator. However, recent research has shown that the central entity can perfectly reconstruct private data from shared model updates by maliciously initializing the model's parameters. In this paper, we propose QBI, a novel bias initialization method that significantly enhances reconstruction capabilities. This is accomplished by directly solving for bias values yielding sparse activation patterns. Further, we propose PAIRS, an algorithm that builds on QBI. PAIRS can be deployed when a separate dataset from the target domain is available to further increase the percentage of data that can be fully recovered. Measured by the percentage of samples that can be perfectly reconstructed from batches of various sizes, our approach achieves significant improvements over previous methods with gains of up to 50% on ImageNet and up to 60% on the IMDB sentiment analysis text dataset. Furthermore, we establish theoretical limits for attacks leveraging stochastic gradient sparsity, providing a foundation for understanding the fundamental constraints of these attacks. We empirically assess these limits using synthetic datasets. Finally, we propose and evaluate AGGP, a defensive framework designed to prevent gradient sparsity attacks, contributing to the development of more secure and private federated learning systems.
A translational pathway of deep learning methods in GastroIntestinal Endoscopy
Oct 12, 2020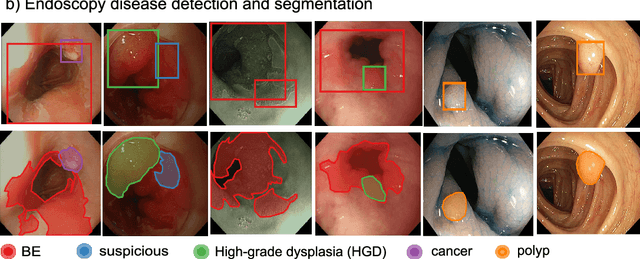
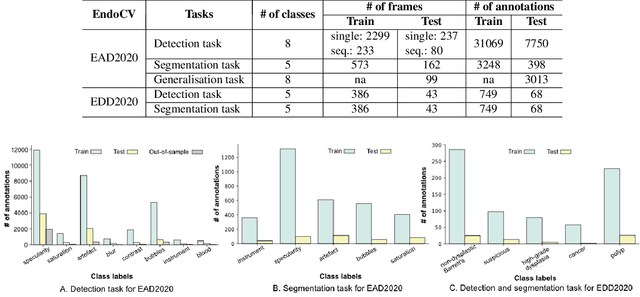
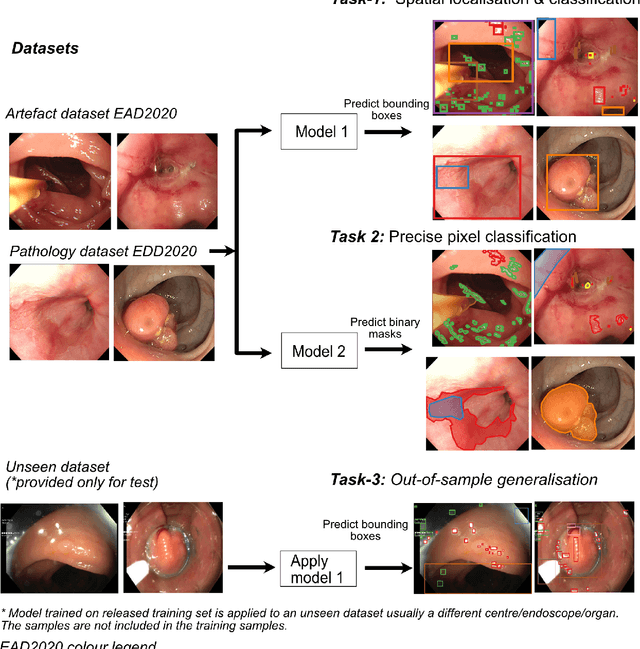
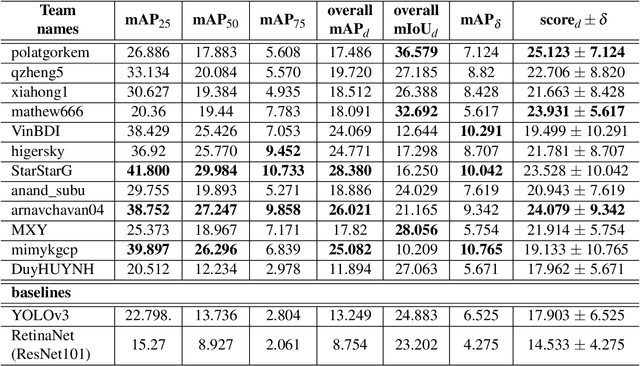
Abstract:The Endoscopy Computer Vision Challenge (EndoCV) is a crowd-sourcing initiative to address eminent problems in developing reliable computer aided detection and diagnosis endoscopy systems and suggest a pathway for clinical translation of technologies. Whilst endoscopy is a widely used diagnostic and treatment tool for hollow-organs, there are several core challenges often faced by endoscopists, mainly: 1) presence of multi-class artefacts that hinder their visual interpretation, and 2) difficulty in identifying subtle precancerous precursors and cancer abnormalities. Artefacts often affect the robustness of deep learning methods applied to the gastrointestinal tract organs as they can be confused with tissue of interest. EndoCV2020 challenges are designed to address research questions in these remits. In this paper, we present a summary of methods developed by the top 17 teams and provide an objective comparison of state-of-the-art methods and methods designed by the participants for two sub-challenges: i) artefact detection and segmentation (EAD2020), and ii) disease detection and segmentation (EDD2020). Multi-center, multi-organ, multi-class, and multi-modal clinical endoscopy datasets were compiled for both EAD2020 and EDD2020 sub-challenges. An out-of-sample generalisation ability of detection algorithms was also evaluated. Whilst most teams focused on accuracy improvements, only a few methods hold credibility for clinical usability. The best performing teams provided solutions to tackle class imbalance, and variabilities in size, origin, modality and occurrences by exploring data augmentation, data fusion, and optimal class thresholding techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge