Ece Isik-Polat
Assessing generalisability of deep learning-based polyp detection and segmentation methods through a computer vision challenge
Feb 24, 2022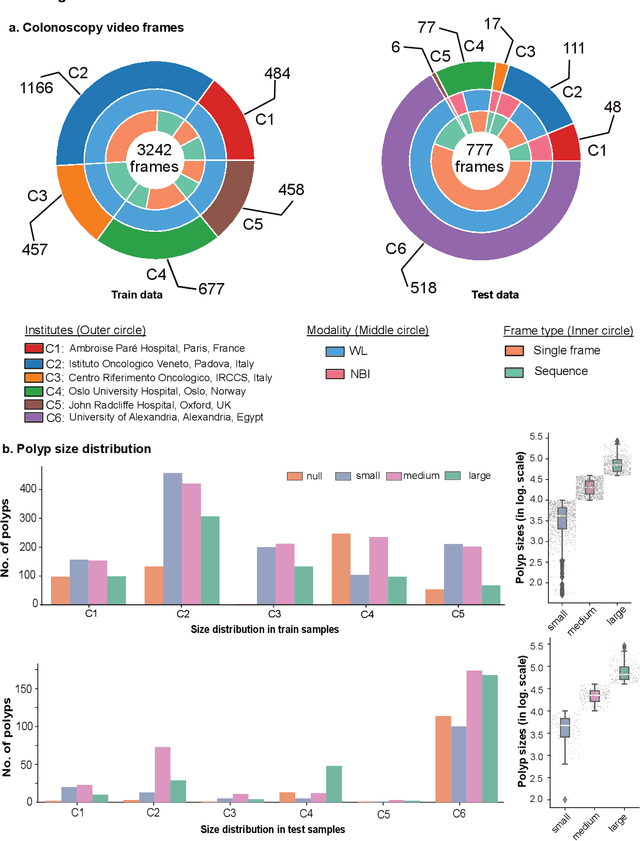
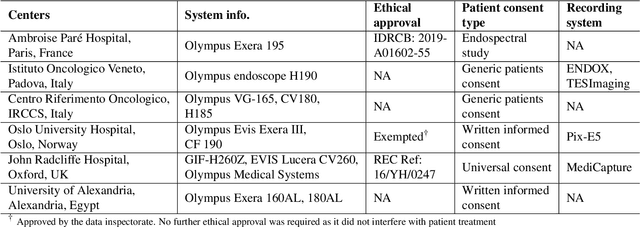
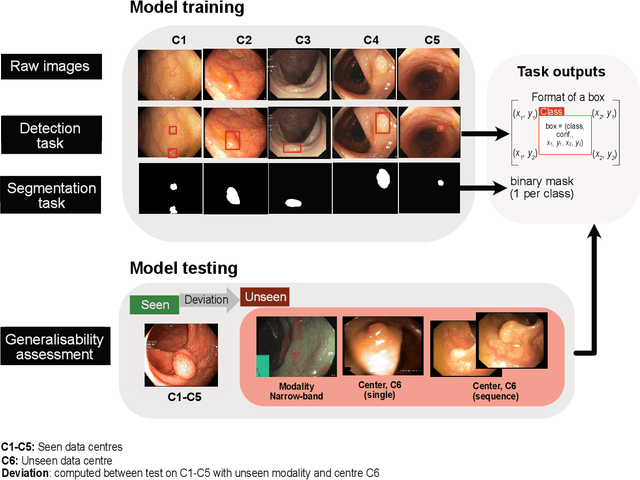
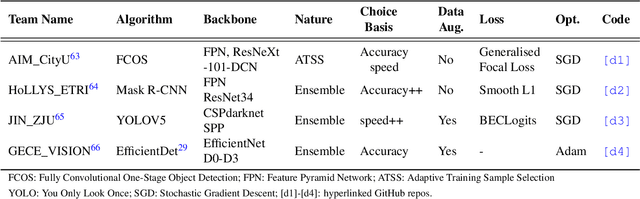
Abstract:Polyps are well-known cancer precursors identified by colonoscopy. However, variability in their size, location, and surface largely affect identification, localisation, and characterisation. Moreover, colonoscopic surveillance and removal of polyps (referred to as polypectomy ) are highly operator-dependent procedures. There exist a high missed detection rate and incomplete removal of colonic polyps due to their variable nature, the difficulties to delineate the abnormality, the high recurrence rates, and the anatomical topography of the colon. There have been several developments in realising automated methods for both detection and segmentation of these polyps using machine learning. However, the major drawback in most of these methods is their ability to generalise to out-of-sample unseen datasets that come from different centres, modalities and acquisition systems. To test this hypothesis rigorously we curated a multi-centre and multi-population dataset acquired from multiple colonoscopy systems and challenged teams comprising machine learning experts to develop robust automated detection and segmentation methods as part of our crowd-sourcing Endoscopic computer vision challenge (EndoCV) 2021. In this paper, we analyse the detection results of the four top (among seven) teams and the segmentation results of the five top teams (among 16). Our analyses demonstrate that the top-ranking teams concentrated on accuracy (i.e., accuracy > 80% on overall Dice score on different validation sets) over real-time performance required for clinical applicability. We further dissect the methods and provide an experiment-based hypothesis that reveals the need for improved generalisability to tackle diversity present in multi-centre datasets.
Evaluation and Analysis of Different Aggregation and Hyperparameter Selection Methods for Federated Brain Tumor Segmentation
Feb 16, 2022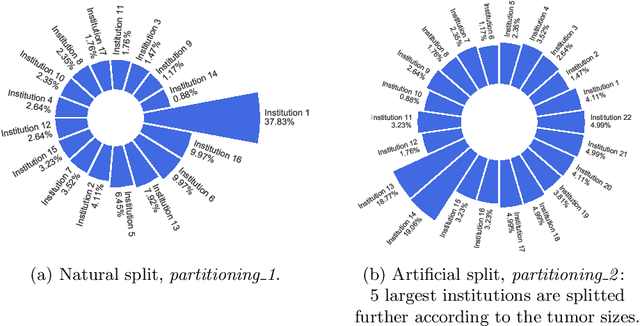
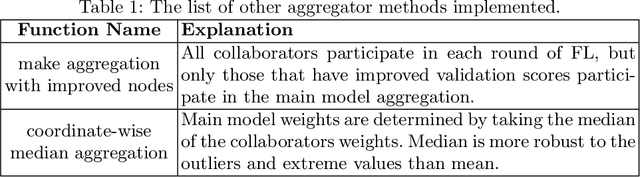

Abstract:Availability of large, diverse, and multi-national datasets is crucial for the development of effective and clinically applicable AI systems in the medical imaging domain. However, forming a global model by bringing these datasets together at a central location, comes along with various data privacy and ownership problems. To alleviate these problems, several recent studies focus on the federated learning paradigm, a distributed learning approach for decentralized data. Federated learning leverages all the available data without any need for sharing collaborators' data with each other or collecting them on a central server. Studies show that federated learning can provide competitive performance with conventional central training, while having a good generalization capability. In this work, we have investigated several federated learning approaches on the brain tumor segmentation problem. We explore different strategies for faster convergence and better performance which can also work on strong Non-IID cases.
BARFED: Byzantine Attack-Resistant Federated Averaging Based on Outlier Elimination
Nov 08, 2021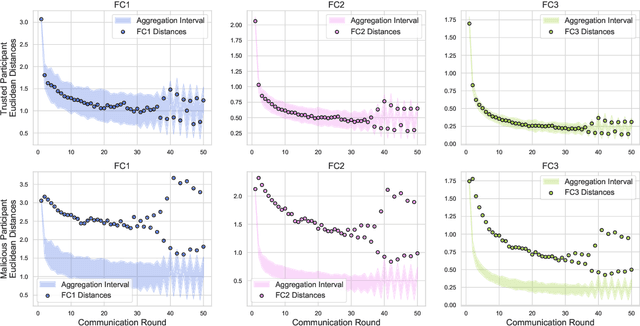
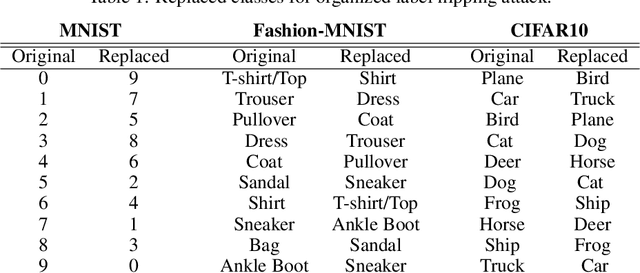
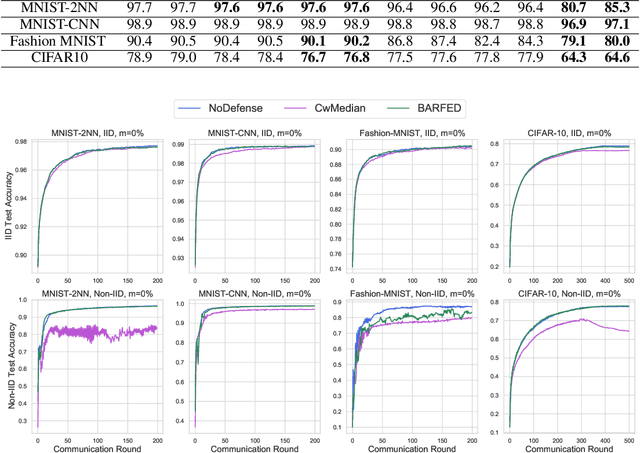
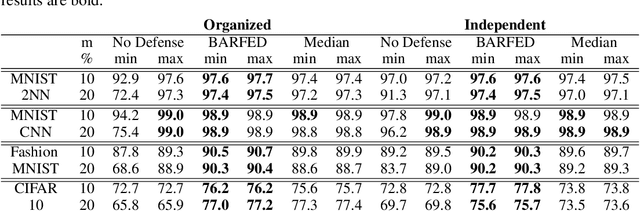
Abstract:In federated learning, each participant trains its local model with its own data and a global model is formed at a trusted server by aggregating model updates coming from these participants. Since the server has no effect and visibility on the training procedure of the participants to ensure privacy, the global model becomes vulnerable to attacks such as data poisoning and model poisoning. Although many defense algorithms have recently been proposed to address these attacks, they often make strong assumptions that do not agree with the nature of federated learning, such as Non-IID datasets. Moreover, they mostly lack comprehensive experimental analyses. In this work, we propose a defense algorithm called BARFED that does not make any assumptions about data distribution, update similarity of participants, or the ratio of the malicious participants. BARFED mainly considers the outlier status of participant updates for each layer of the model architecture based on the distance to the global model. Hence, the participants that do not have any outlier layer are involved in model aggregation. We perform extensive experiments on many grounds and show that the proposed approach provides a robust defense against different attacks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge