Soumya Gupta
Patch-level instance-group discrimination with pretext-invariant learning for colitis scoring
Jul 11, 2022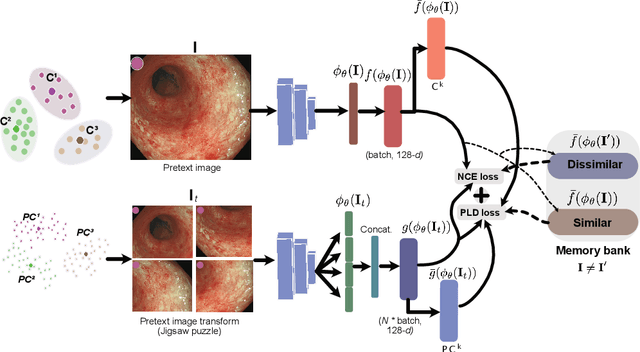
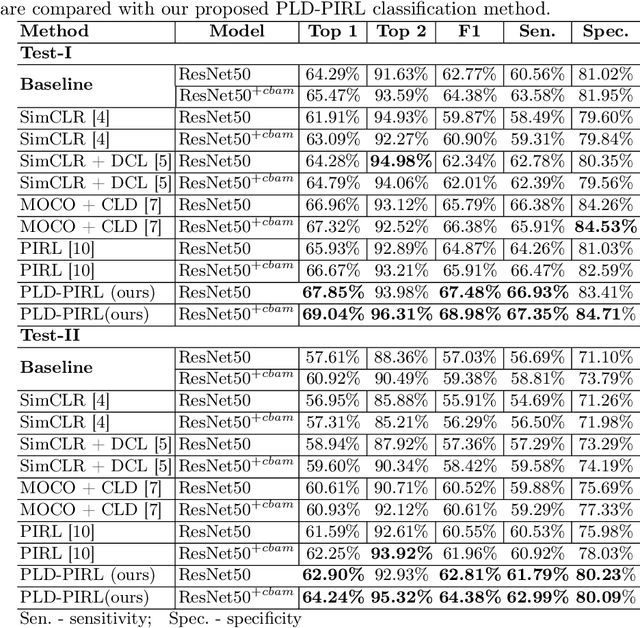
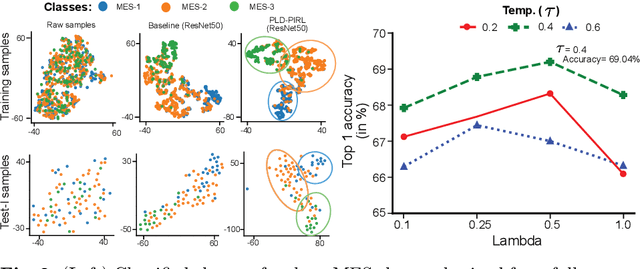
Abstract:Inflammatory bowel disease (IBD), in particular ulcerative colitis (UC), is graded by endoscopists and this assessment is the basis for risk stratification and therapy monitoring. Presently, endoscopic characterisation is largely operator dependant leading to sometimes undesirable clinical outcomes for patients with IBD. We focus on the Mayo Endoscopic Scoring (MES) system which is widely used but requires the reliable identification of subtle changes in mucosal inflammation. Most existing deep learning classification methods cannot detect these fine-grained changes which make UC grading such a challenging task. In this work, we introduce a novel patch-level instance-group discrimination with pretext-invariant representation learning (PLD-PIRL) for self-supervised learning (SSL). Our experiments demonstrate both improved accuracy and robustness compared to the baseline supervised network and several state-of-the-art SSL methods. Compared to the baseline (ResNet50) supervised classification our proposed PLD-PIRL obtained an improvement of 4.75% on hold-out test data and 6.64% on unseen center test data for top-1 accuracy.
EndoUDA: A modality independent segmentation approach for endoscopy imaging
Jul 12, 2021
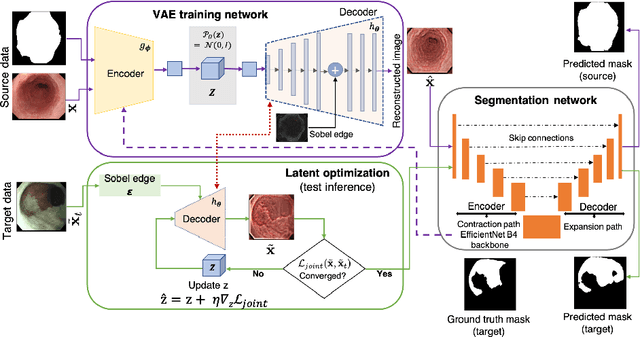
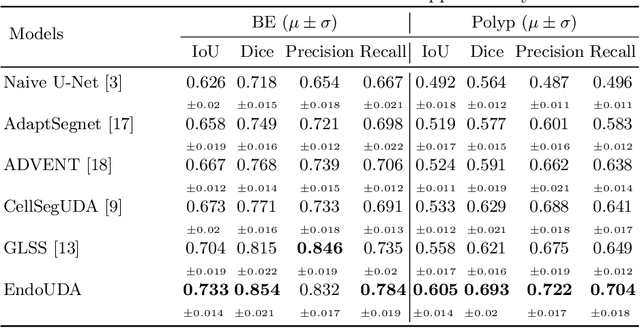
Abstract:Gastrointestinal (GI) cancer precursors require frequent monitoring for risk stratification of patients. Automated segmentation methods can help to assess risk areas more accurately, and assist in therapeutic procedures or even removal. In clinical practice, addition to the conventional white-light imaging (WLI), complimentary modalities such as narrow-band imaging (NBI) and fluorescence imaging are used. While, today most segmentation approaches are supervised and only concentrated on a single modality dataset, this work exploits to use a target-independent unsupervised domain adaptation (UDA) technique that is capable to generalize to an unseen target modality. In this context, we propose a novel UDA-based segmentation method that couples the variational autoencoder and U-Net with a common EfficientNet-B4 backbone, and uses a joint loss for latent-space optimization for target samples. We show that our model can generalize to unseen target NBI (target) modality when trained using only WLI (source) modality. Our experiments on both upper and lower GI endoscopy data show the effectiveness of our approach compared to naive supervised approach and state-of-the-art UDA segmentation methods.
Multi-class motion-based semantic segmentation for ureteroscopy and laser lithotripsy
Apr 02, 2021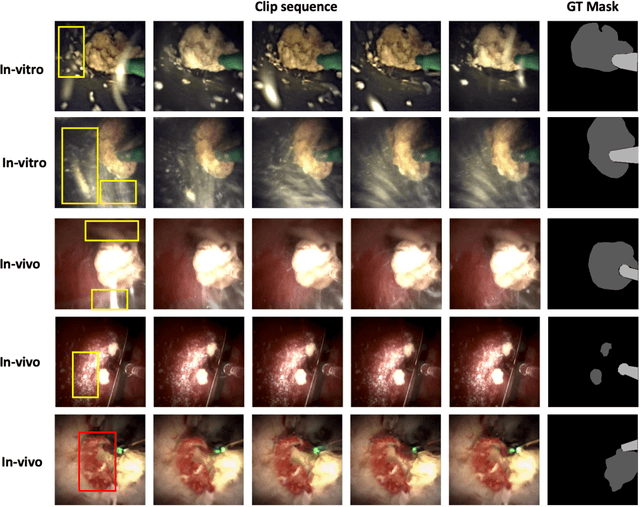
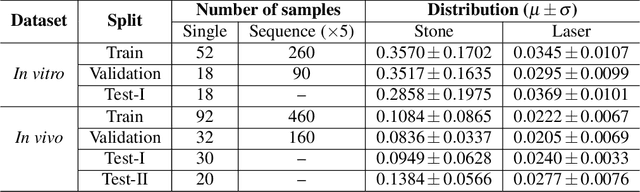
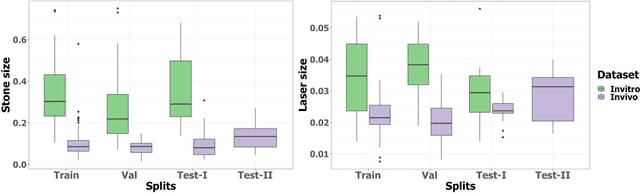
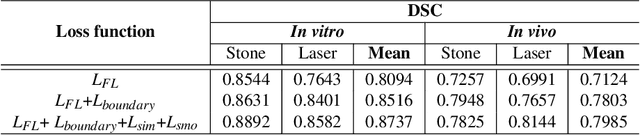
Abstract:Kidney stones represent a considerable burden for public health-care systems. Ureteroscopy with laser lithotripsy has evolved as the most commonly used technique for the treatment of kidney stones. Automated segmentation of kidney stones and laser fiber is an important initial step to performing any automated quantitative analysis of the stones, particularly stone-size estimation, that helps the surgeon decide if the stone requires more fragmentation. Factors such as turbid fluid inside the cavity, specularities, motion blur due to kidney movements and camera motion, bleeding, and stone debris impact the quality of vision within the kidney and lead to extended operative times. To the best of our knowledge, this is the first attempt made towards multi-class segmentation in ureteroscopy and laser lithotripsy data. We propose an end-to-end CNN-based framework for the segmentation of stones and laser fiber. The proposed approach utilizes two sub-networks: HybResUNet, a version of residual U-Net, that uses residual connections in the encoder path of U-Net and a DVFNet that generates DVF predictions which are then used to prune the prediction maps. We also present ablation studies that combine dilated convolutions, recurrent and residual connections, ASPP and attention gate. We propose a compound loss function that improves our segmentation performance. We have also provided an ablation study to determine the optimal data augmentation strategy. Our qualitative and quantitative results illustrate that our proposed method outperforms SOTA methods such as UNet and DeepLabv3+ showing an improvement of 5.2% and 15.93%, respectively, for the combined mean of DSC and JI in our invivo test dataset. We also show that our proposed model generalizes better on a new clinical dataset showing a mean improvement of 25.4%, 20%, and 11% over UNet, HybResUNet, and DeepLabv3+, respectively, for the same metric.
Unsupervised Adversarial Domain Adaptation For Barrett's Segmentation
Dec 09, 2020

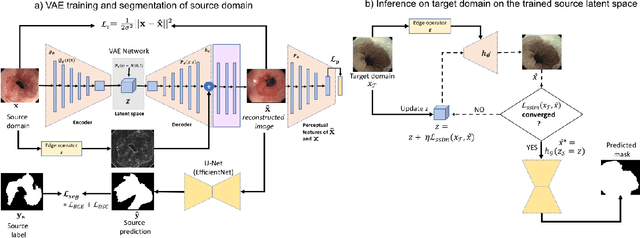
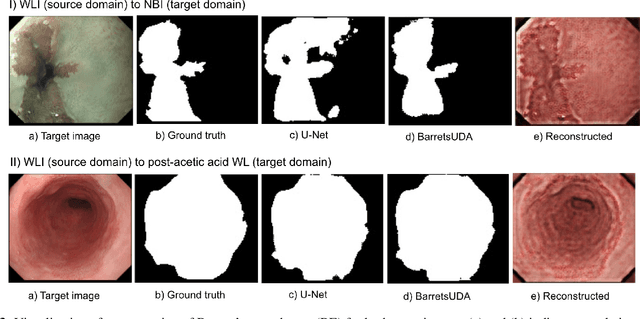
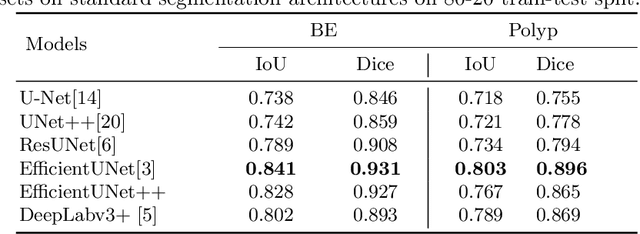
Abstract:Barrett's oesophagus (BE) is one of the early indicators of esophageal cancer. Patients with BE are monitored and undergo ablation therapies to minimise the risk, thereby making it eminent to identify the BE area precisely. Automated segmentation can help clinical endoscopists to assess and treat BE area more accurately. Endoscopy imaging of BE can include multiple modalities in addition to the conventional white light (WL) modality. Supervised models require large amount of manual annotations incorporating all data variability in the training data. However, it becomes cumbersome, tedious and labour intensive work to generate manual annotations, and additionally modality specific expertise is required. In this work, we aim to alleviate this problem by applying an unsupervised domain adaptation technique (UDA). Here, UDA is trained on white light endoscopy images as source domain and are well-adapted to generalise to produce segmentation on different imaging modalities as target domain, namely narrow band imaging and post acetic-acid WL imaging. Our dataset consists of a total of 871 images consisting of both source and target domains. Our results show that the UDA-based approach outperforms traditional supervised U-Net segmentation by nearly 10% on both Dice similarity coefficient and intersection-over-union.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge