Unsupervised Adversarial Domain Adaptation For Barrett's Segmentation
Paper and Code
Dec 09, 2020

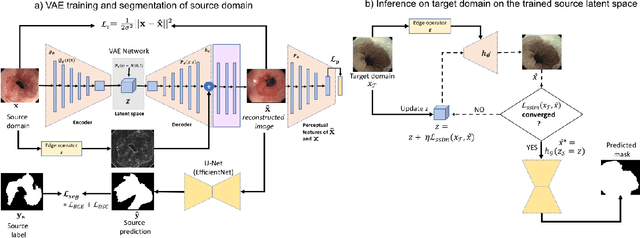
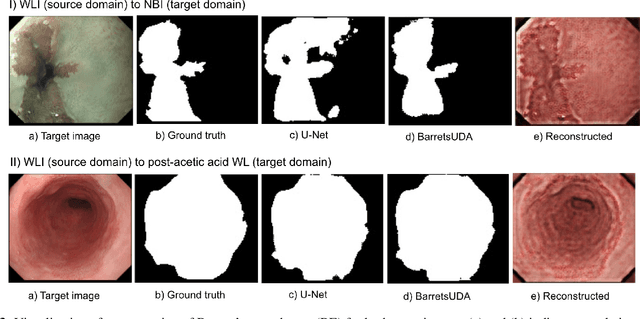
Barrett's oesophagus (BE) is one of the early indicators of esophageal cancer. Patients with BE are monitored and undergo ablation therapies to minimise the risk, thereby making it eminent to identify the BE area precisely. Automated segmentation can help clinical endoscopists to assess and treat BE area more accurately. Endoscopy imaging of BE can include multiple modalities in addition to the conventional white light (WL) modality. Supervised models require large amount of manual annotations incorporating all data variability in the training data. However, it becomes cumbersome, tedious and labour intensive work to generate manual annotations, and additionally modality specific expertise is required. In this work, we aim to alleviate this problem by applying an unsupervised domain adaptation technique (UDA). Here, UDA is trained on white light endoscopy images as source domain and are well-adapted to generalise to produce segmentation on different imaging modalities as target domain, namely narrow band imaging and post acetic-acid WL imaging. Our dataset consists of a total of 871 images consisting of both source and target domains. Our results show that the UDA-based approach outperforms traditional supervised U-Net segmentation by nearly 10% on both Dice similarity coefficient and intersection-over-union.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge