Osama E. Salem
Assessing generalisability of deep learning-based polyp detection and segmentation methods through a computer vision challenge
Feb 24, 2022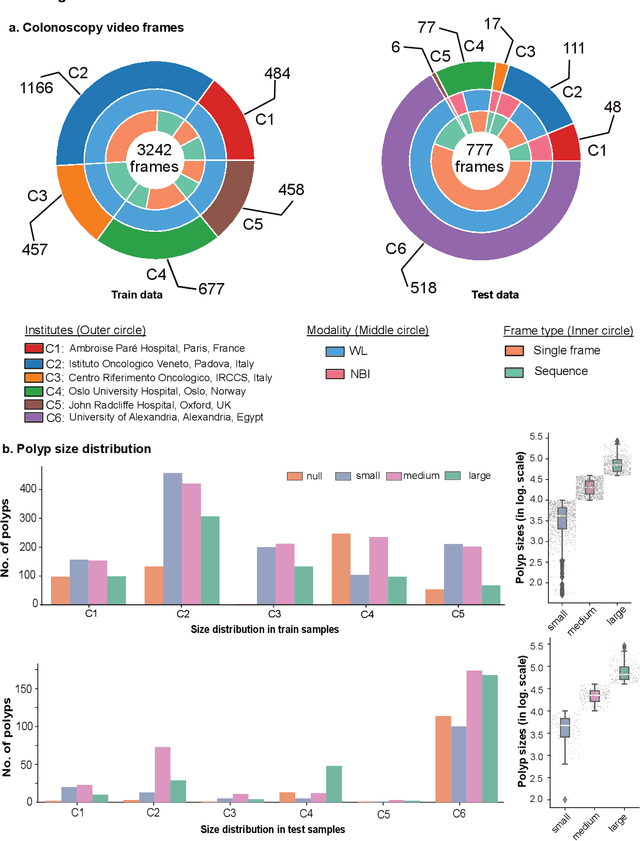
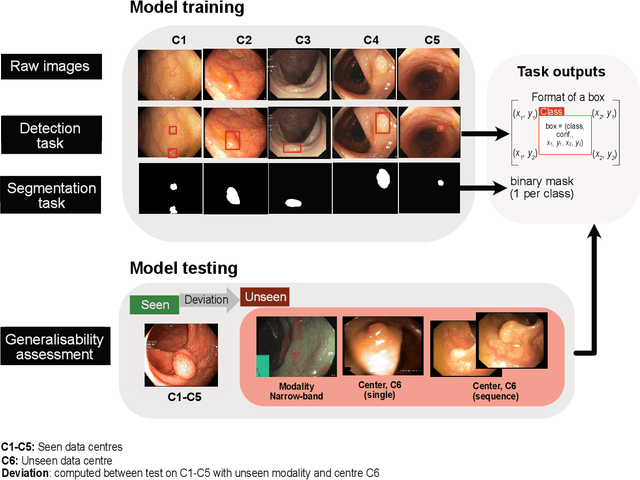
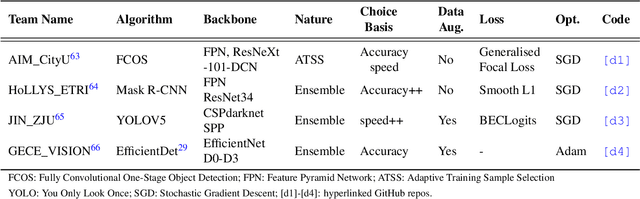
Abstract:Polyps are well-known cancer precursors identified by colonoscopy. However, variability in their size, location, and surface largely affect identification, localisation, and characterisation. Moreover, colonoscopic surveillance and removal of polyps (referred to as polypectomy ) are highly operator-dependent procedures. There exist a high missed detection rate and incomplete removal of colonic polyps due to their variable nature, the difficulties to delineate the abnormality, the high recurrence rates, and the anatomical topography of the colon. There have been several developments in realising automated methods for both detection and segmentation of these polyps using machine learning. However, the major drawback in most of these methods is their ability to generalise to out-of-sample unseen datasets that come from different centres, modalities and acquisition systems. To test this hypothesis rigorously we curated a multi-centre and multi-population dataset acquired from multiple colonoscopy systems and challenged teams comprising machine learning experts to develop robust automated detection and segmentation methods as part of our crowd-sourcing Endoscopic computer vision challenge (EndoCV) 2021. In this paper, we analyse the detection results of the four top (among seven) teams and the segmentation results of the five top teams (among 16). Our analyses demonstrate that the top-ranking teams concentrated on accuracy (i.e., accuracy > 80% on overall Dice score on different validation sets) over real-time performance required for clinical applicability. We further dissect the methods and provide an experiment-based hypothesis that reveals the need for improved generalisability to tackle diversity present in multi-centre datasets.
PolypGen: A multi-center polyp detection and segmentation dataset for generalisability assessment
Jun 08, 2021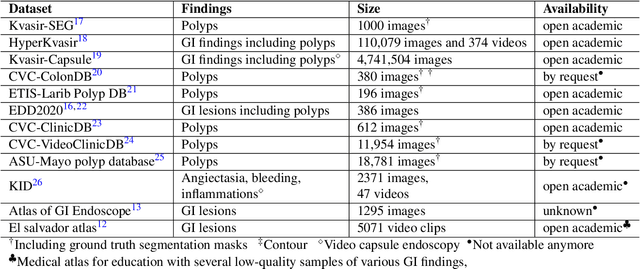
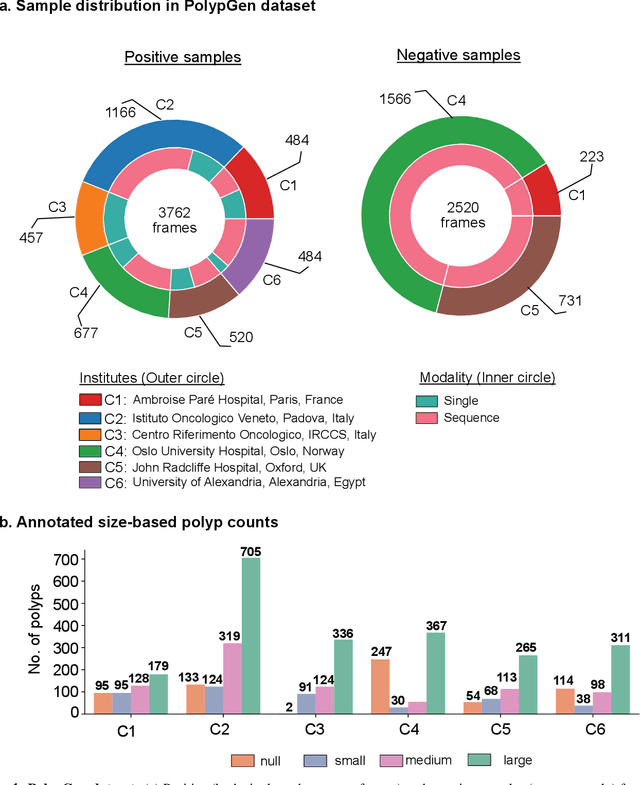
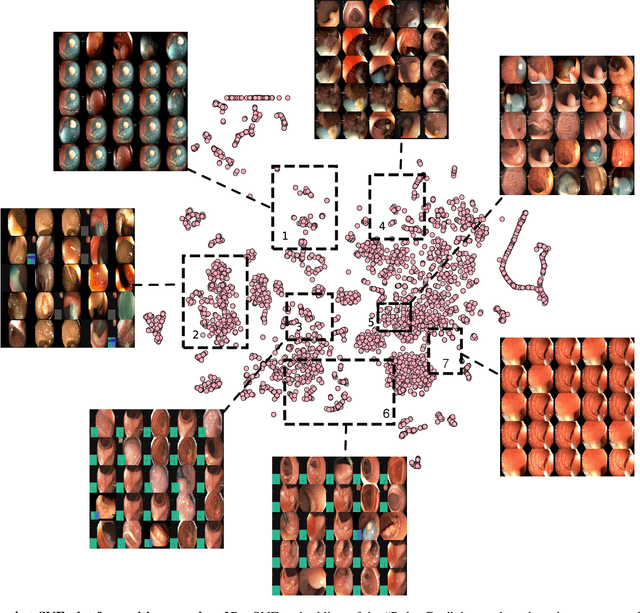
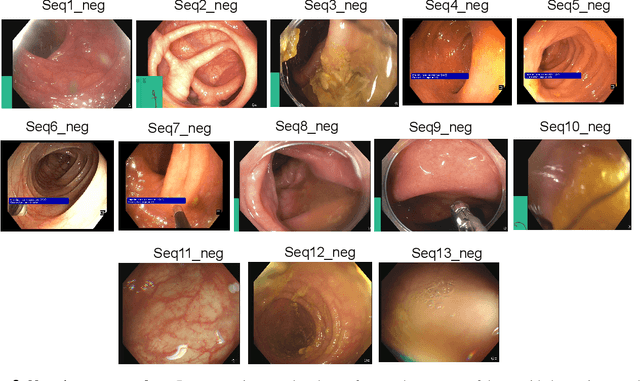
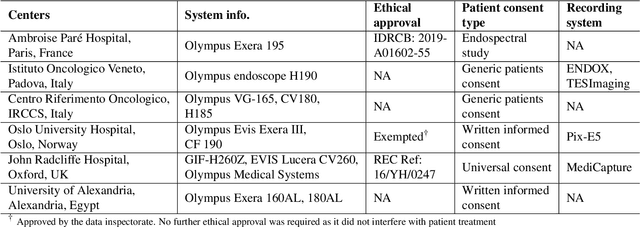
Abstract:Polyps in the colon are widely known as cancer precursors identified by colonoscopy either related to diagnostic work-up for symptoms, colorectal cancer screening or systematic surveillance of certain diseases. Whilst most polyps are benign, the number, size and the surface structure of the polyp are tightly linked to the risk of colon cancer. There exists a high missed detection rate and incomplete removal of colon polyps due to the variable nature, difficulties to delineate the abnormality, high recurrence rates and the anatomical topography of the colon. In the past, several methods have been built to automate polyp detection and segmentation. However, the key issue of most methods is that they have not been tested rigorously on a large multi-center purpose-built dataset. Thus, these methods may not generalise to different population datasets as they overfit to a specific population and endoscopic surveillance. To this extent, we have curated a dataset from 6 different centers incorporating more than 300 patients. The dataset includes both single frame and sequence data with 3446 annotated polyp labels with precise delineation of polyp boundaries verified by six senior gastroenterologists. To our knowledge, this is the most comprehensive detection and pixel-level segmentation dataset curated by a team of computational scientists and expert gastroenterologists. This dataset has been originated as the part of the Endocv2021 challenge aimed at addressing generalisability in polyp detection and segmentation. In this paper, we provide comprehensive insight into data construction and annotation strategies, annotation quality assurance and technical validation for our extended EndoCV2021 dataset which we refer to as PolypGen.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge