Dominique Lamarque
Deep learning-based image exposure enhancement as a pre-processing for an accurate 3D colon surface reconstruction
Apr 14, 2023



Abstract:This contribution shows how an appropriate image pre-processing can improve a deep-learning based 3D reconstruction of colon parts. The assumption is that, rather than global image illumination corrections, local under- and over-exposures should be corrected in colonoscopy. An overview of the pipeline including the image exposure correction and a RNN-SLAM is first given. Then, this paper quantifies the reconstruction accuracy of the endoscope trajectory in the colon with and without appropriate illumination correction
Multi-Scale Structural-aware Exposure Correction for Endoscopic Imaging
Oct 26, 2022



Abstract:Endoscopy is the most widely used imaging technique for the diagnosis of cancerous lesions in hollow organs. However, endoscopic images are often affected by illumination artefacts: image parts may be over- or underexposed according to the light source pose and the tissue orientation. These artifacts have a strong negative impact on the performance of computer vision or AI-based diagnosis tools. Although endoscopic image enhancement methods are greatly required, little effort has been devoted to over- and under-exposition enhancement in real-time. This contribution presents an extension to the objective function of LMSPEC, a method originally introduced to enhance images from natural scenes. It is used here for the exposure correction in endoscopic imaging and the preservation of structural information. To the best of our knowledge, this contribution is the first one that addresses the enhancement of endoscopic images using deep learning (DL) methods. Tested on the Endo4IE dataset, the proposed implementation has yielded a significant improvement over LMSPEC reaching a SSIM increase of 4.40% and 4.21% for over- and underexposed images, respectively.
A Novel Hybrid Endoscopic Dataset for Evaluating Machine Learning-based Photometric Image Enhancement Models
Jul 06, 2022
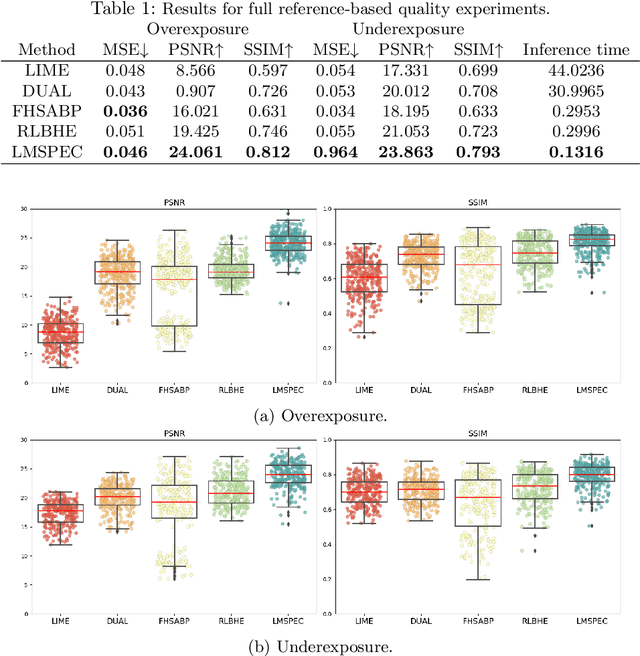
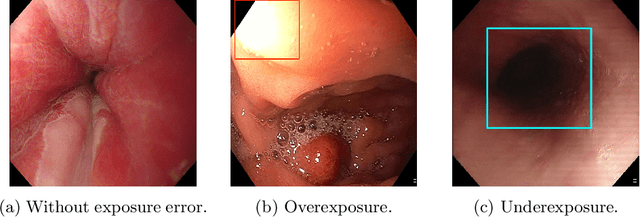
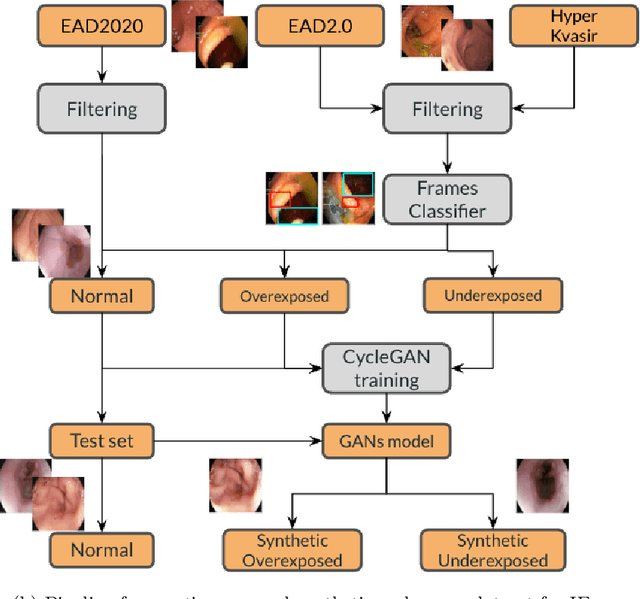
Abstract:Endoscopy is the most widely used medical technique for cancer and polyp detection inside hollow organs. However, images acquired by an endoscope are frequently affected by illumination artefacts due to the enlightenment source orientation. There exist two major issues when the endoscope's light source pose suddenly changes: overexposed and underexposed tissue areas are produced. These two scenarios can result in misdiagnosis due to the lack of information in the affected zones or hamper the performance of various computer vision methods (e.g., SLAM, structure from motion, optical flow) used during the non invasive examination. The aim of this work is two-fold: i) to introduce a new synthetically generated data-set generated by a generative adversarial techniques and ii) and to explore both shallow based and deep learning-based image-enhancement methods in overexposed and underexposed lighting conditions. Best quantitative results (i.e., metric based results), were obtained by the deep-learnnig-based LMSPEC method,besides a running time around 7.6 fps)
Evaluating object detector ensembles for improving the robustness of artifact detection in endoscopic video streams
Jun 15, 2022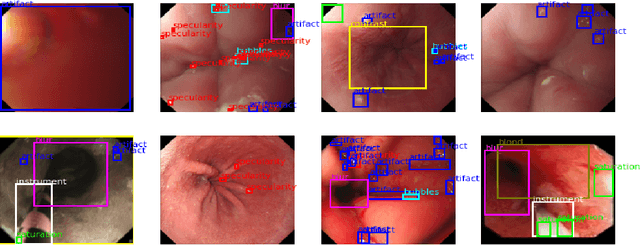
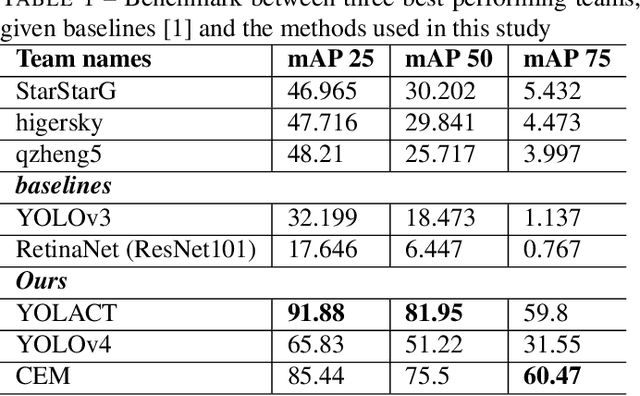
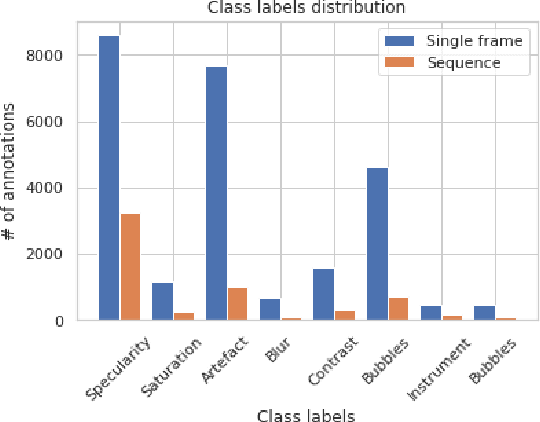
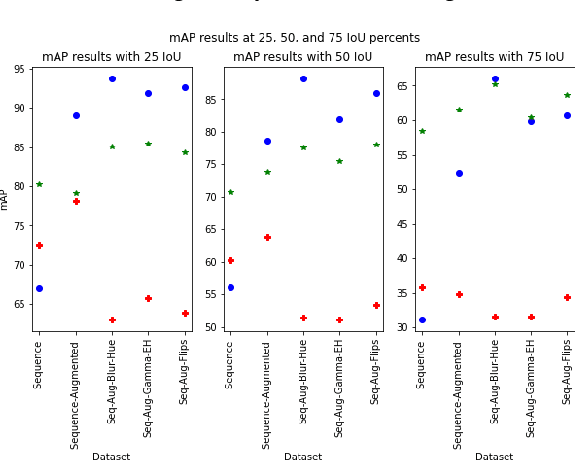
Abstract:In this contribution we use an ensemble deep-learning method for combining the prediction of two individual one-stage detectors (i.e., YOLOv4 and Yolact) with the aim to detect artefacts in endoscopic images. This ensemble strategy enabled us to improve the robustness of the individual models without harming their real-time computation capabilities. We demonstrated the effectiveness of our approach by training and testing the two individual models and various ensemble configurations on the "Endoscopic Artifact Detection Challenge" dataset. Extensive experiments show the superiority, in terms of mean average precision, of the ensemble approach over the individual models and previous works in the state of the art.
Assessing generalisability of deep learning-based polyp detection and segmentation methods through a computer vision challenge
Feb 24, 2022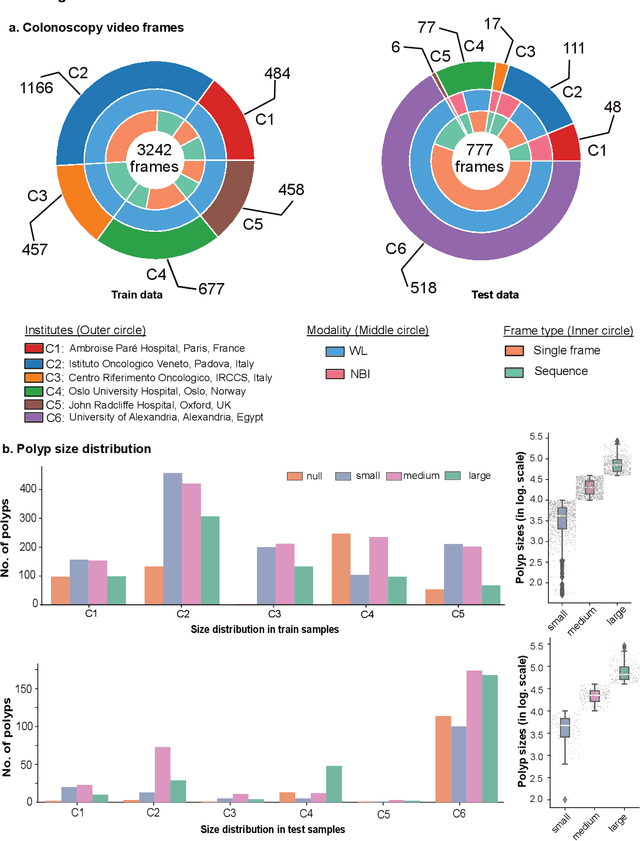
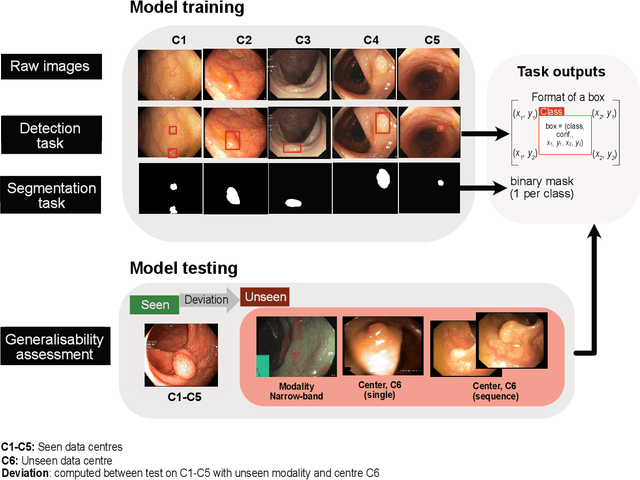
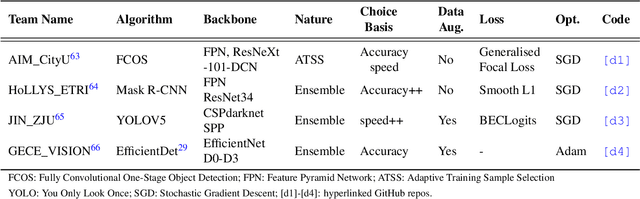
Abstract:Polyps are well-known cancer precursors identified by colonoscopy. However, variability in their size, location, and surface largely affect identification, localisation, and characterisation. Moreover, colonoscopic surveillance and removal of polyps (referred to as polypectomy ) are highly operator-dependent procedures. There exist a high missed detection rate and incomplete removal of colonic polyps due to their variable nature, the difficulties to delineate the abnormality, the high recurrence rates, and the anatomical topography of the colon. There have been several developments in realising automated methods for both detection and segmentation of these polyps using machine learning. However, the major drawback in most of these methods is their ability to generalise to out-of-sample unseen datasets that come from different centres, modalities and acquisition systems. To test this hypothesis rigorously we curated a multi-centre and multi-population dataset acquired from multiple colonoscopy systems and challenged teams comprising machine learning experts to develop robust automated detection and segmentation methods as part of our crowd-sourcing Endoscopic computer vision challenge (EndoCV) 2021. In this paper, we analyse the detection results of the four top (among seven) teams and the segmentation results of the five top teams (among 16). Our analyses demonstrate that the top-ranking teams concentrated on accuracy (i.e., accuracy > 80% on overall Dice score on different validation sets) over real-time performance required for clinical applicability. We further dissect the methods and provide an experiment-based hypothesis that reveals the need for improved generalisability to tackle diversity present in multi-centre datasets.
PolypGen: A multi-center polyp detection and segmentation dataset for generalisability assessment
Jun 08, 2021
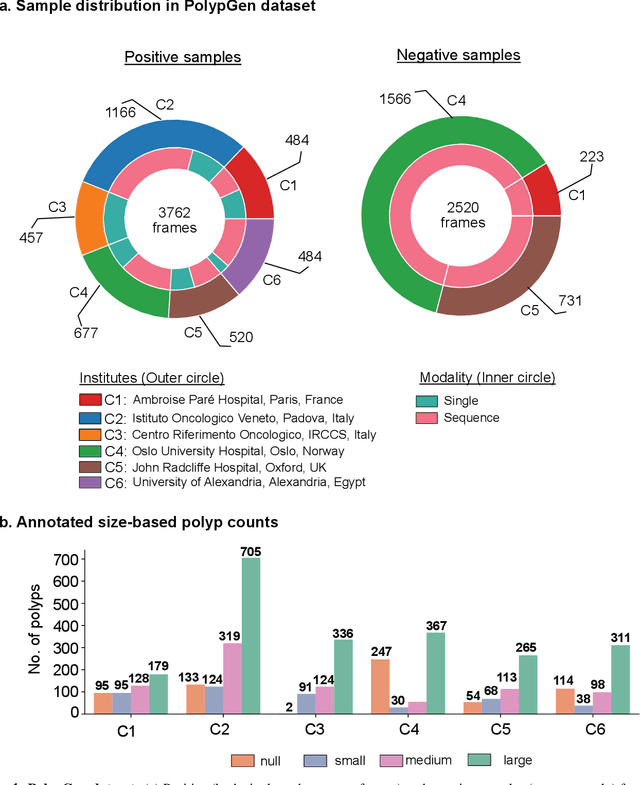
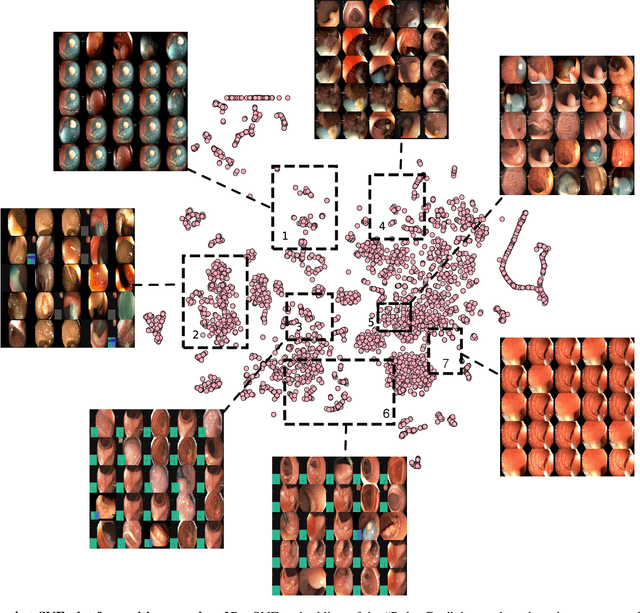
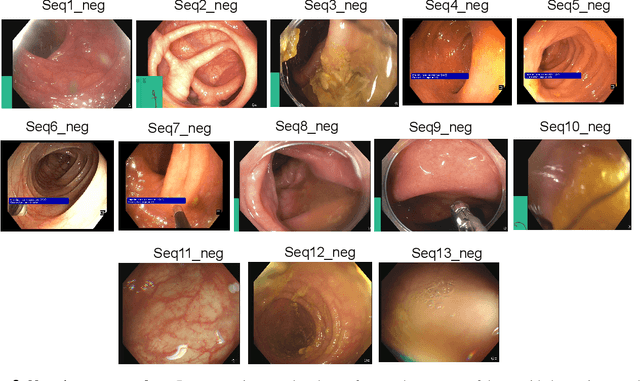
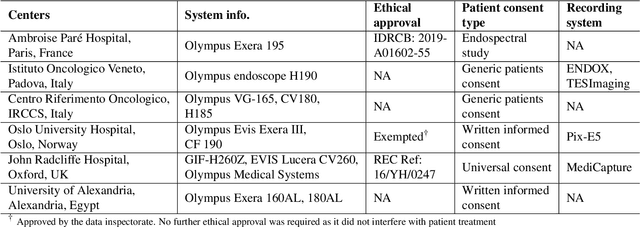
Abstract:Polyps in the colon are widely known as cancer precursors identified by colonoscopy either related to diagnostic work-up for symptoms, colorectal cancer screening or systematic surveillance of certain diseases. Whilst most polyps are benign, the number, size and the surface structure of the polyp are tightly linked to the risk of colon cancer. There exists a high missed detection rate and incomplete removal of colon polyps due to the variable nature, difficulties to delineate the abnormality, high recurrence rates and the anatomical topography of the colon. In the past, several methods have been built to automate polyp detection and segmentation. However, the key issue of most methods is that they have not been tested rigorously on a large multi-center purpose-built dataset. Thus, these methods may not generalise to different population datasets as they overfit to a specific population and endoscopic surveillance. To this extent, we have curated a dataset from 6 different centers incorporating more than 300 patients. The dataset includes both single frame and sequence data with 3446 annotated polyp labels with precise delineation of polyp boundaries verified by six senior gastroenterologists. To our knowledge, this is the most comprehensive detection and pixel-level segmentation dataset curated by a team of computational scientists and expert gastroenterologists. This dataset has been originated as the part of the Endocv2021 challenge aimed at addressing generalisability in polyp detection and segmentation. In this paper, we provide comprehensive insight into data construction and annotation strategies, annotation quality assurance and technical validation for our extended EndoCV2021 dataset which we refer to as PolypGen.
A translational pathway of deep learning methods in GastroIntestinal Endoscopy
Oct 12, 2020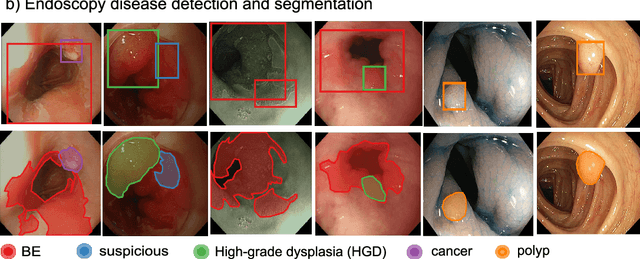
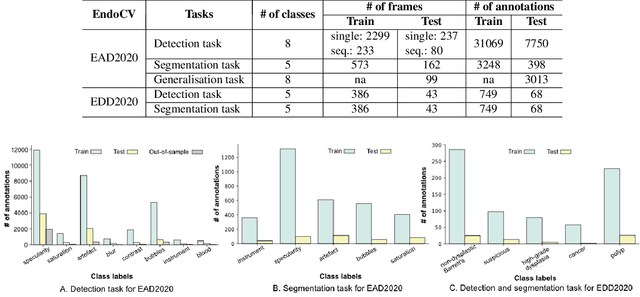
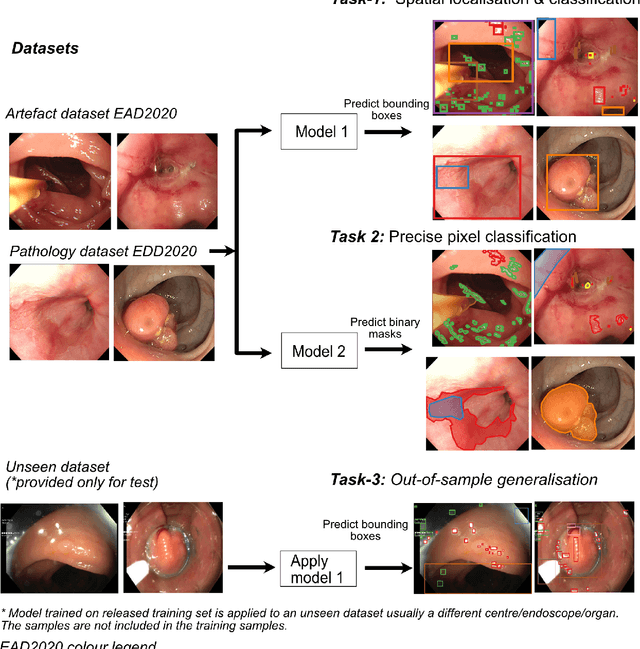
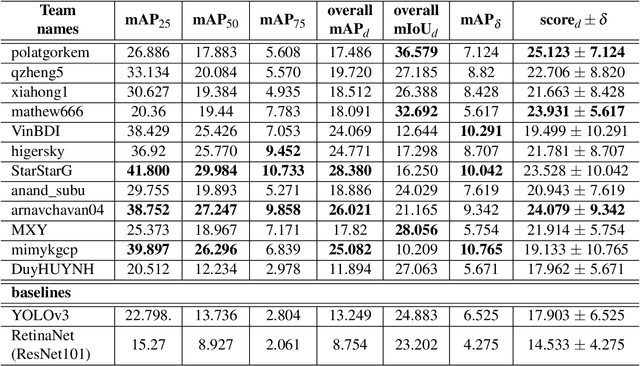
Abstract:The Endoscopy Computer Vision Challenge (EndoCV) is a crowd-sourcing initiative to address eminent problems in developing reliable computer aided detection and diagnosis endoscopy systems and suggest a pathway for clinical translation of technologies. Whilst endoscopy is a widely used diagnostic and treatment tool for hollow-organs, there are several core challenges often faced by endoscopists, mainly: 1) presence of multi-class artefacts that hinder their visual interpretation, and 2) difficulty in identifying subtle precancerous precursors and cancer abnormalities. Artefacts often affect the robustness of deep learning methods applied to the gastrointestinal tract organs as they can be confused with tissue of interest. EndoCV2020 challenges are designed to address research questions in these remits. In this paper, we present a summary of methods developed by the top 17 teams and provide an objective comparison of state-of-the-art methods and methods designed by the participants for two sub-challenges: i) artefact detection and segmentation (EAD2020), and ii) disease detection and segmentation (EDD2020). Multi-center, multi-organ, multi-class, and multi-modal clinical endoscopy datasets were compiled for both EAD2020 and EDD2020 sub-challenges. An out-of-sample generalisation ability of detection algorithms was also evaluated. Whilst most teams focused on accuracy improvements, only a few methods hold credibility for clinical usability. The best performing teams provided solutions to tackle class imbalance, and variabilities in size, origin, modality and occurrences by exploring data augmentation, data fusion, and optimal class thresholding techniques.
Endoscopy disease detection challenge 2020
Mar 07, 2020
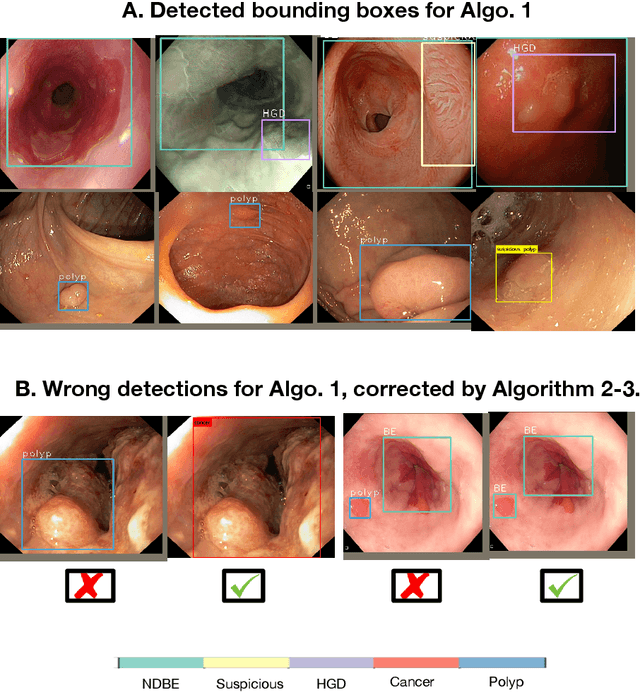
Abstract:Whilst many technologies are built around endoscopy, there is a need to have a comprehensive dataset collected from multiple centers to address the generalization issues with most deep learning frameworks. What could be more important than disease detection and localization? Through our extensive network of clinical and computational experts, we have collected, curated and annotated gastrointestinal endoscopy video frames. We have released this dataset and have launched disease detection and segmentation challenge EDD2020 https://edd2020.grand-challenge.org to address the limitations and explore new directions. EDD2020 is a crowd sourcing initiative to test the feasibility of recent deep learning methods and to promote research for building robust technologies. In this paper, we provide an overview of the EDD2020 dataset, challenge tasks, evaluation strategies and a short summary of results on test data. A detailed paper will be drafted after the challenge workshop with more detailed analysis of the results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge