Clare Verrill
Histology-informed tiling of whole tissue sections improves the interpretability and predictability of cancer relapse and genetic alterations
Nov 13, 2025
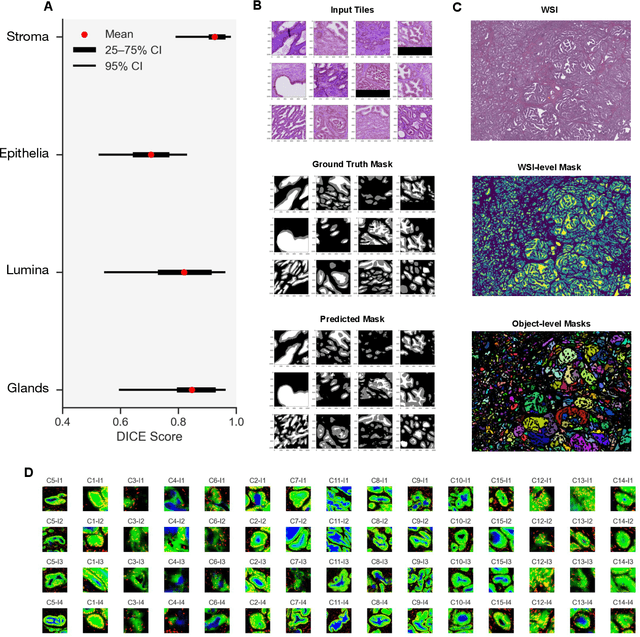
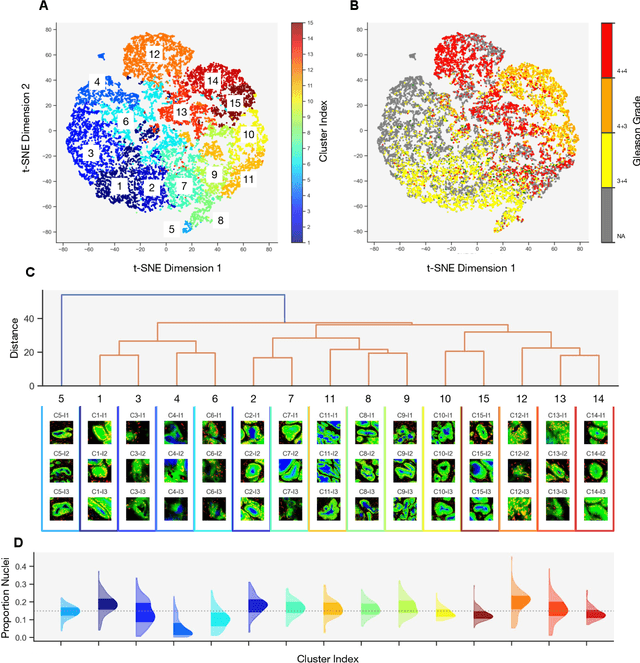
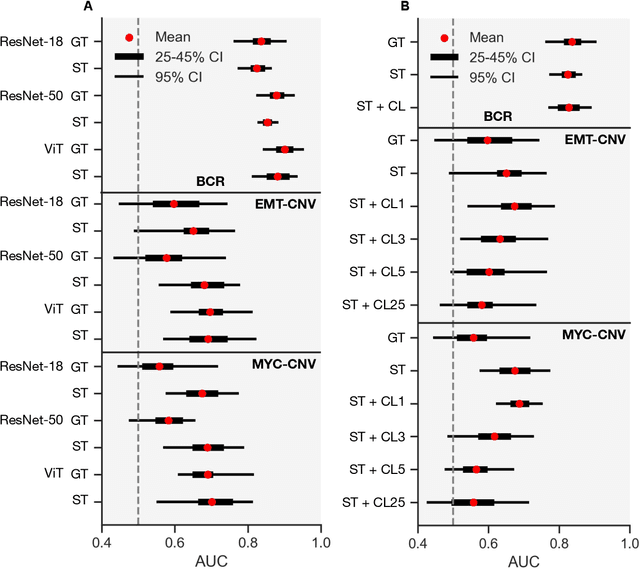
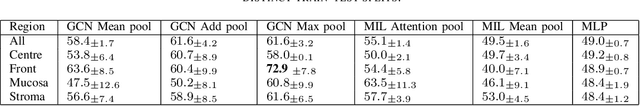
Abstract:Histopathologists establish cancer grade by assessing histological structures, such as glands in prostate cancer. Yet, digital pathology pipelines often rely on grid-based tiling that ignores tissue architecture. This introduces irrelevant information and limits interpretability. We introduce histology-informed tiling (HIT), which uses semantic segmentation to extract glands from whole slide images (WSIs) as biologically meaningful input patches for multiple-instance learning (MIL) and phenotyping. Trained on 137 samples from the ProMPT cohort, HIT achieved a gland-level Dice score of 0.83 +/- 0.17. By extracting 380,000 glands from 760 WSIs across ICGC-C and TCGA-PRAD cohorts, HIT improved MIL models AUCs by 10% for detecting copy number variation (CNVs) in genes related to epithelial-mesenchymal transitions (EMT) and MYC, and revealed 15 gland clusters, several of which were associated with cancer relapse, oncogenic mutations, and high Gleason. Therefore, HIT improved the accuracy and interpretability of MIL predictions, while streamlining computations by focussing on biologically meaningful structures during feature extraction.
Beyond attention: deriving biologically interpretable insights from weakly-supervised multiple-instance learning models
Sep 07, 2023



Abstract:Recent advances in attention-based multiple instance learning (MIL) have improved our insights into the tissue regions that models rely on to make predictions in digital pathology. However, the interpretability of these approaches is still limited. In particular, they do not report whether high-attention regions are positively or negatively associated with the class labels or how well these regions correspond to previously established clinical and biological knowledge. We address this by introducing a post-training methodology to analyse MIL models. Firstly, we introduce prediction-attention-weighted (PAW) maps by combining tile-level attention and prediction scores produced by a refined encoder, allowing us to quantify the predictive contribution of high-attention regions. Secondly, we introduce a biological feature instantiation technique by integrating PAW maps with nuclei segmentation masks. This further improves interpretability by providing biologically meaningful features related to the cellular organisation of the tissue and facilitates comparisons with known clinical features. We illustrate the utility of our approach by comparing PAW maps obtained for prostate cancer diagnosis (i.e. samples containing malignant tissue, 381/516 tissue samples) and prognosis (i.e. samples from patients with biochemical recurrence following surgery, 98/663 tissue samples) in a cohort of patients from the international cancer genome consortium (ICGC UK Prostate Group). Our approach reveals that regions that are predictive of adverse prognosis do not tend to co-locate with the tumour regions, indicating that non-cancer cells should also be studied when evaluating prognosis.
A Graph Based Neural Network Approach to Immune Profiling of Multiplexed Tissue Samples
Feb 01, 2022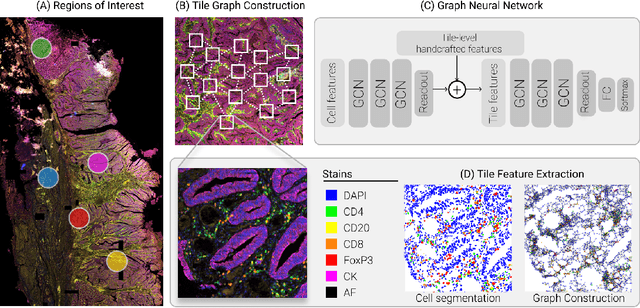
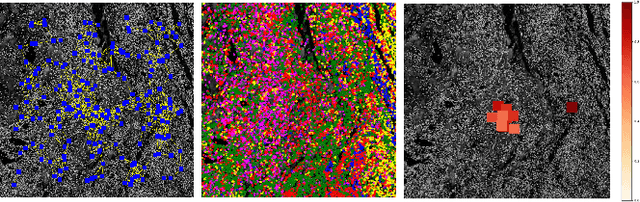
Abstract:Multiplexed immunofluorescence provides an unprecedented opportunity for studying specific cell-to-cell and cell microenvironment interactions. We employ graph neural networks to combine features obtained from tissue morphology with measurements of protein expression to profile the tumour microenvironment associated with different tumour stages. Our framework presents a new approach to analysing and processing these complex multi-dimensional datasets that overcomes some of the key challenges in analysing these data and opens up the opportunity to abstract biologically meaningful interactions.
Ink removal from histopathology whole slide images by combining classification, detection and image generation models
May 10, 2019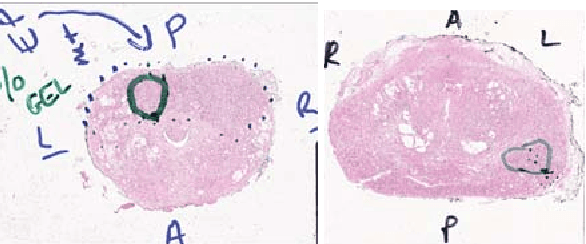
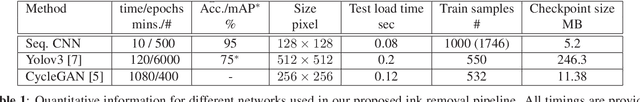

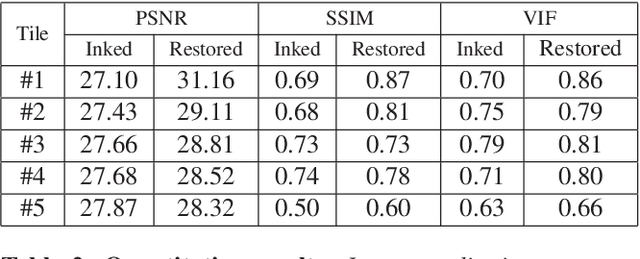
Abstract:Histopathology slides are routinely marked by pathologists using permanent ink markers that should not be removed as they form part of the medical record. Often tumour regions are marked up for the purpose of highlighting features or other downstream processing such an gene sequencing. Once digitised there is no established method for removing this information from the whole slide images limiting its usability in research and study. Removal of marker ink from these high-resolution whole slide images is non-trivial and complex problem as they contaminate different regions and in an inconsistent manner. We propose an efficient pipeline using convolution neural networks that results in ink-free images without compromising information and image resolution. Our pipeline includes a sequential classical convolution neural network for accurate classification of contaminated image tiles, a fast region detector and a domain adaptive cycle consistent adversarial generative model for restoration of foreground pixels. Both quantitative and qualitative results on four different whole slide images show that our approach yields visually coherent ink-free whole slide images.
Improving Whole Slide Segmentation Through Visual Context - A Systematic Study
Jun 11, 2018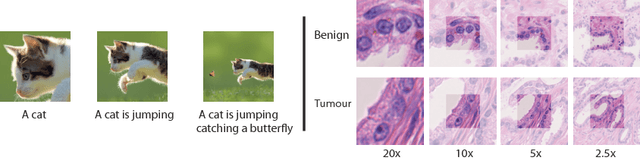
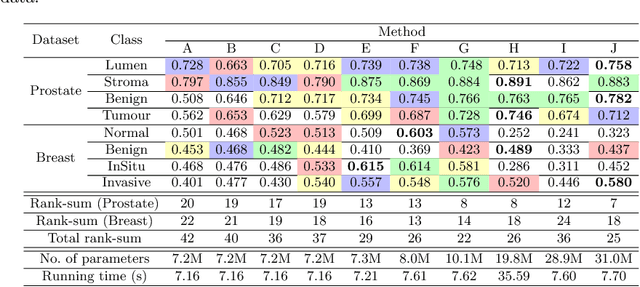
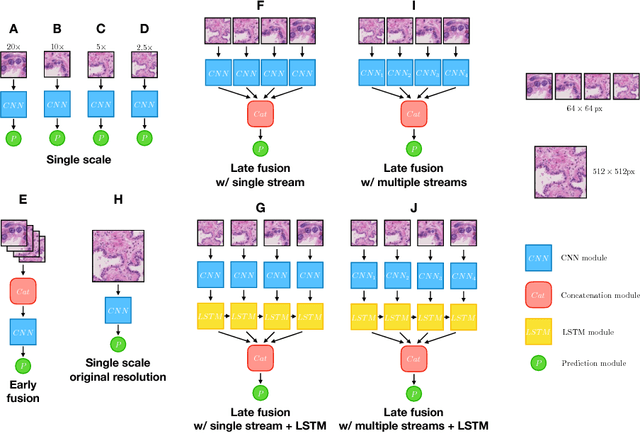
Abstract:While challenging, the dense segmentation of histology images is a necessary first step to assess changes in tissue architecture and cellular morphology. Although specific convolutional neural network architectures have been applied with great success to the problem, few effectively incorporate visual context information from multiple scales. With this paper, we present a systematic comparison of different architectures to assess how including multi-scale information affects segmentation performance. A publicly available breast cancer and a locally collected prostate cancer datasets are being utilised for this study. The results support our hypothesis that visual context and scale play a crucial role in histology image classification problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge