Yilong Yang
GRPO Privacy Is at Risk: A Membership Inference Attack Against Reinforcement Learning With Verifiable Rewards
Nov 18, 2025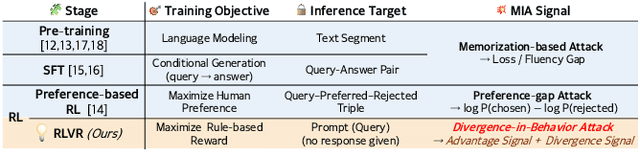
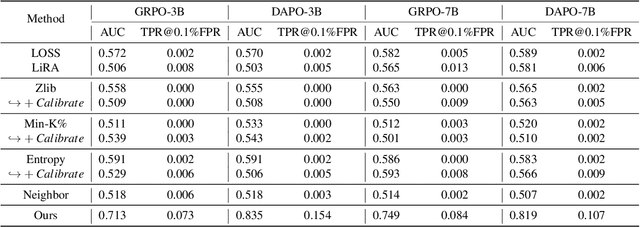
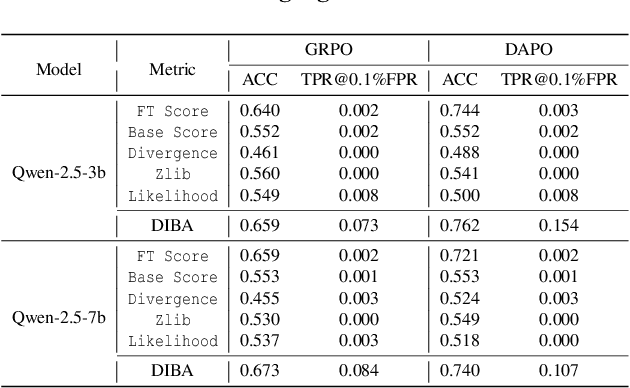
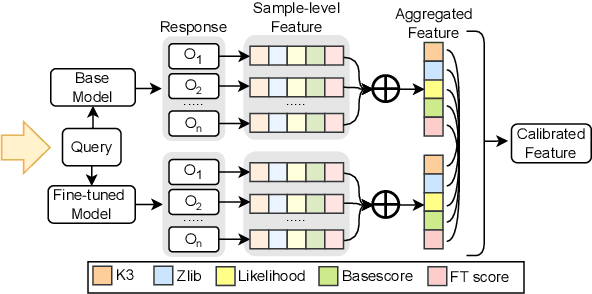
Abstract:Membership inference attacks (MIAs) on large language models (LLMs) pose significant privacy risks across various stages of model training. Recent advances in Reinforcement Learning with Verifiable Rewards (RLVR) have brought a profound paradigm shift in LLM training, particularly for complex reasoning tasks. However, the on-policy nature of RLVR introduces a unique privacy leakage pattern: since training relies on self-generated responses without fixed ground-truth outputs, membership inference must now determine whether a given prompt (independent of any specific response) is used during fine-tuning. This creates a threat where leakage arises not from answer memorization. To audit this novel privacy risk, we propose Divergence-in-Behavior Attack (DIBA), the first membership inference framework specifically designed for RLVR. DIBA shifts the focus from memorization to behavioral change, leveraging measurable shifts in model behavior across two axes: advantage-side improvement (e.g., correctness gain) and logit-side divergence (e.g., policy drift). Through comprehensive evaluations, we demonstrate that DIBA significantly outperforms existing baselines, achieving around 0.8 AUC and an order-of-magnitude higher TPR@0.1%FPR. We validate DIBA's superiority across multiple settings--including in-distribution, cross-dataset, cross-algorithm, black-box scenarios, and extensions to vision-language models. Furthermore, our attack remains robust under moderate defensive measures. To the best of our knowledge, this is the first work to systematically analyze privacy vulnerabilities in RLVR, revealing that even in the absence of explicit supervision, training data exposure can be reliably inferred through behavioral traces.
Improving Sustainability of Adversarial Examples in Class-Incremental Learning
Nov 12, 2025



Abstract:Current adversarial examples (AEs) are typically designed for static models. However, with the wide application of Class-Incremental Learning (CIL), models are no longer static and need to be updated with new data distributed and labeled differently from the old ones. As a result, existing AEs often fail after CIL updates due to significant domain drift. In this paper, we propose SAE to enhance the sustainability of AEs against CIL. The core idea of SAE is to enhance the robustness of AE semantics against domain drift by making them more similar to the target class while distinguishing them from all other classes. Achieving this is challenging, as relying solely on the initial CIL model to optimize AE semantics often leads to overfitting. To resolve the problem, we propose a Semantic Correction Module. This module encourages the AE semantics to be generalized, based on a visual-language model capable of producing universal semantics. Additionally, it incorporates the CIL model to correct the optimization direction of the AE semantics, guiding them closer to the target class. To further reduce fluctuations in AE semantics, we propose a Filtering-and-Augmentation Module, which first identifies non-target examples with target-class semantics in the latent space and then augments them to foster more stable semantics. Comprehensive experiments demonstrate that SAE outperforms baselines by an average of 31.28% when updated with a 9-fold increase in the number of classes.
NICE: Improving Panoptic Narrative Detection and Segmentation with Cascading Collaborative Learning
Oct 23, 2023Abstract:Panoptic Narrative Detection (PND) and Segmentation (PNS) are two challenging tasks that involve identifying and locating multiple targets in an image according to a long narrative description. In this paper, we propose a unified and effective framework called NICE that can jointly learn these two panoptic narrative recognition tasks. Existing visual grounding tasks use a two-branch paradigm, but applying this directly to PND and PNS can result in prediction conflict due to their intrinsic many-to-many alignment property. To address this, we introduce two cascading modules based on the barycenter of the mask, which are Coordinate Guided Aggregation (CGA) and Barycenter Driven Localization (BDL), responsible for segmentation and detection, respectively. By linking PNS and PND in series with the barycenter of segmentation as the anchor, our approach naturally aligns the two tasks and allows them to complement each other for improved performance. Specifically, CGA provides the barycenter as a reference for detection, reducing BDL's reliance on a large number of candidate boxes. BDL leverages its excellent properties to distinguish different instances, which improves the performance of CGA for segmentation. Extensive experiments demonstrate that NICE surpasses all existing methods by a large margin, achieving 4.1% for PND and 2.9% for PNS over the state-of-the-art. These results validate the effectiveness of our proposed collaborative learning strategy. The project of this work is made publicly available at https://github.com/Mr-Neko/NICE.
3D Shape-Based Myocardial Infarction Prediction Using Point Cloud Classification Networks
Jul 14, 2023



Abstract:Myocardial infarction (MI) is one of the most prevalent cardiovascular diseases with associated clinical decision-making typically based on single-valued imaging biomarkers. However, such metrics only approximate the complex 3D structure and physiology of the heart and hence hinder a better understanding and prediction of MI outcomes. In this work, we investigate the utility of complete 3D cardiac shapes in the form of point clouds for an improved detection of MI events. To this end, we propose a fully automatic multi-step pipeline consisting of a 3D cardiac surface reconstruction step followed by a point cloud classification network. Our method utilizes recent advances in geometric deep learning on point clouds to enable direct and efficient multi-scale learning on high-resolution surface models of the cardiac anatomy. We evaluate our approach on 1068 UK Biobank subjects for the tasks of prevalent MI detection and incident MI prediction and find improvements of ~13% and ~5% respectively over clinical benchmarks. Furthermore, we analyze the role of each ventricle and cardiac phase for 3D shape-based MI detection and conduct a visual analysis of the morphological and physiological patterns typically associated with MI outcomes.
Rotation-Scale Equivariant Steerable Filters
Apr 10, 2023Abstract:Incorporating either rotation equivariance or scale equivariance into CNNs has proved to be effective in improving models' generalization performance. However, jointly integrating rotation and scale equivariance into CNNs has not been widely explored. Digital histology imaging of biopsy tissue can be captured at arbitrary orientation and magnification and stored at different resolutions, resulting in cells appearing in different scales. When conventional CNNs are applied to histopathology image analysis, the generalization performance of models is limited because 1) a part of the parameters of filters are trained to fit rotation transformation, thus decreasing the capability of learning other discriminative features; 2) fixed-size filters trained on images at a given scale fail to generalize to those at different scales. To deal with these issues, we propose the Rotation-Scale Equivariant Steerable Filter (RSESF), which incorporates steerable filters and scale-space theory. The RSESF contains copies of filters that are linear combinations of Gaussian filters, whose direction is controlled by directional derivatives and whose scale parameters are trainable but constrained to span disjoint scales in successive layers of the network. Extensive experiments on two gland segmentation datasets demonstrate that our method outperforms other approaches, with much fewer trainable parameters and fewer GPU resources required. The source code is available at: https://github.com/ynulonger/RSESF.
Scale-Equivariant UNet for Histopathology Image Segmentation
Apr 10, 2023Abstract:Digital histopathology slides are scanned and viewed under different magnifications and stored as images at different resolutions. Convolutional Neural Networks (CNNs) trained on such images at a given scale fail to generalise to those at different scales. This inability is often addressed by augmenting training data with re-scaled images, allowing a model with sufficient capacity to learn the requisite patterns. Alternatively, designing CNN filters to be scale-equivariant frees up model capacity to learn discriminative features. In this paper, we propose the Scale-Equivariant UNet (SEUNet) for image segmentation by building on scale-space theory. The SEUNet contains groups of filters that are linear combinations of Gaussian basis filters, whose scale parameters are trainable but constrained to span disjoint scales through the layers of the network. Extensive experiments on a nuclei segmentation dataset and a tissue type segmentation dataset demonstrate that our method outperforms other approaches, with much fewer trainable parameters.
ADS_UNet: A Nested UNet for Histopathology Image Segmentation
Apr 10, 2023Abstract:The UNet model consists of fully convolutional network (FCN) layers arranged as contracting encoder and upsampling decoder maps. Nested arrangements of these encoder and decoder maps give rise to extensions of the UNet model, such as UNete and UNet++. Other refinements include constraining the outputs of the convolutional layers to discriminate between segment labels when trained end to end, a property called deep supervision. This reduces feature diversity in these nested UNet models despite their large parameter space. Furthermore, for texture segmentation, pixel correlations at multiple scales contribute to the classification task; hence, explicit deep supervision of shallower layers is likely to enhance performance. In this paper, we propose ADS UNet, a stage-wise additive training algorithm that incorporates resource-efficient deep supervision in shallower layers and takes performance-weighted combinations of the sub-UNets to create the segmentation model. We provide empirical evidence on three histopathology datasets to support the claim that the proposed ADS UNet reduces correlations between constituent features and improves performance while being more resource efficient. We demonstrate that ADS_UNet outperforms state-of-the-art Transformer-based models by 1.08 and 0.6 points on CRAG and BCSS datasets, and yet requires only 37% of GPU consumption and 34% of training time as that required by Transformers.
Cloud-based Federated Boosting for Mobile Crowdsensing
May 09, 2020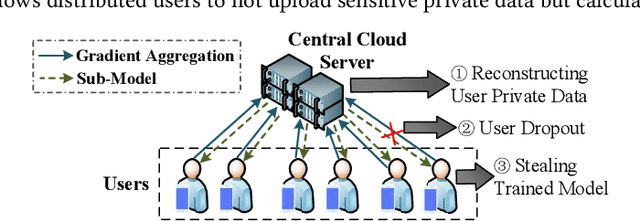
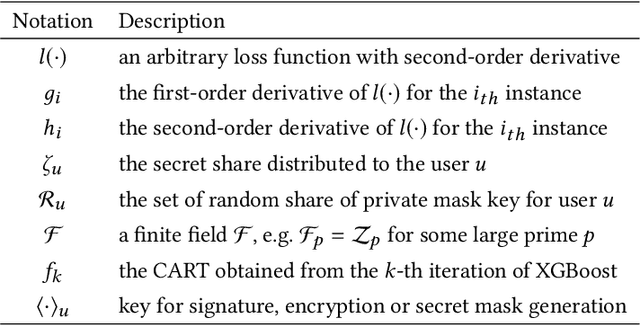
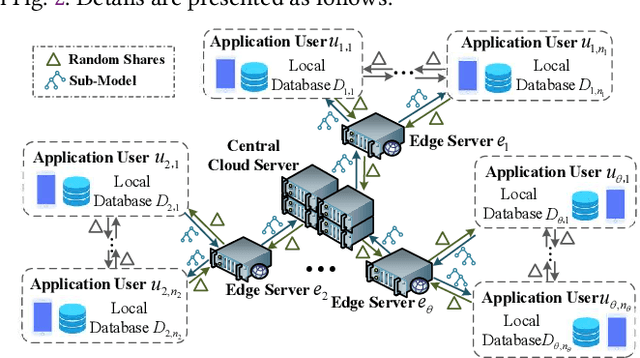
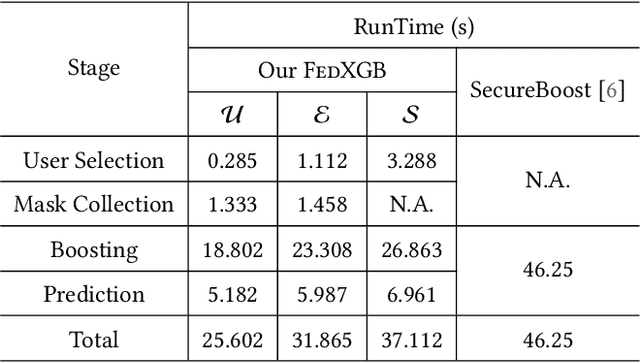
Abstract:The application of federated extreme gradient boosting to mobile crowdsensing apps brings several benefits, in particular high performance on efficiency and classification. However, it also brings a new challenge for data and model privacy protection. Besides it being vulnerable to Generative Adversarial Network (GAN) based user data reconstruction attack, there is not the existing architecture that considers how to preserve model privacy. In this paper, we propose a secret sharing based federated learning architecture FedXGB to achieve the privacy-preserving extreme gradient boosting for mobile crowdsensing. Specifically, we first build a secure classification and regression tree (CART) of XGBoost using secret sharing. Then, we propose a secure prediction protocol to protect the model privacy of XGBoost in mobile crowdsensing. We conduct a comprehensive theoretical analysis and extensive experiments to evaluate the security, effectiveness, and efficiency of FedXGB. The results indicate that FedXGB is secure against the honest-but-curious adversaries and attains less than 1% accuracy loss compared with the original XGBoost model.
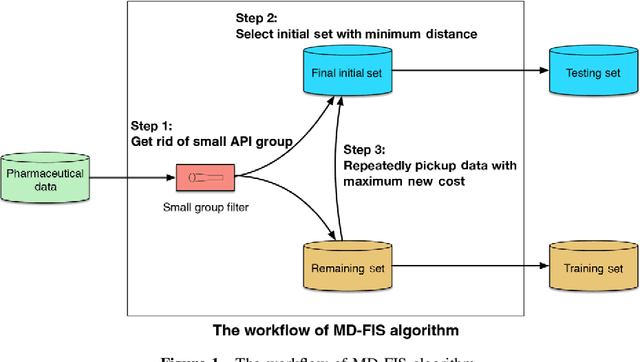
An Integrated Transfer Learning and Multitask Learning Approach for Pharmacokinetic Parameter Prediction
Dec 21, 2018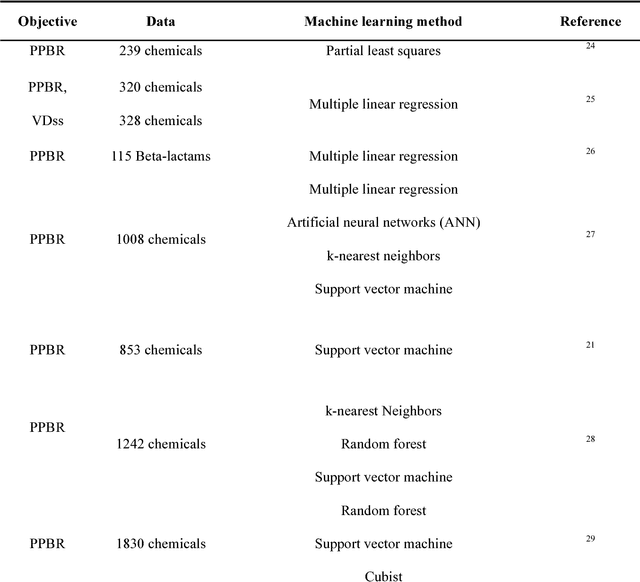
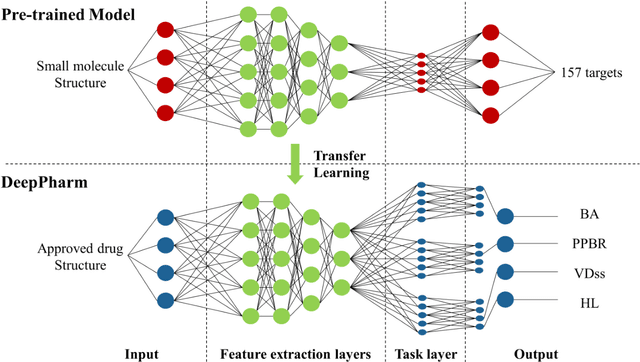
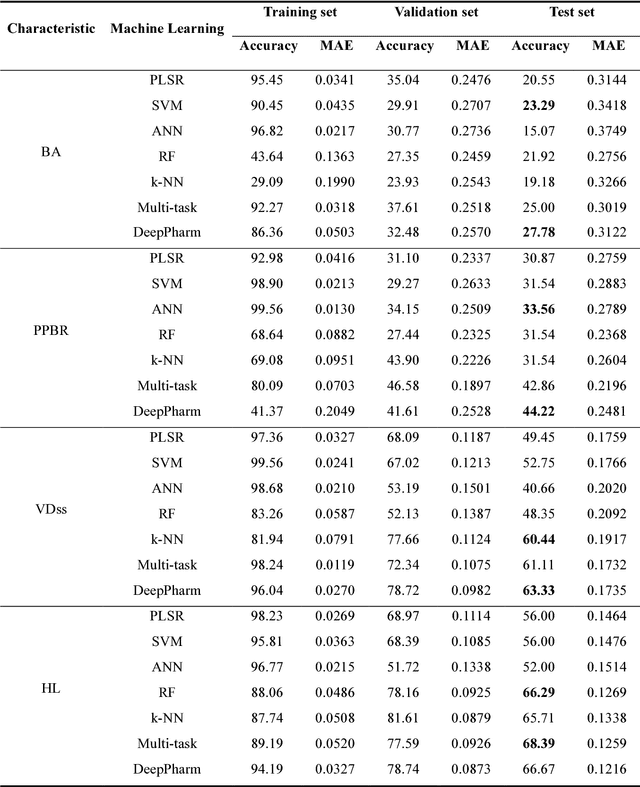

Abstract:Background: Pharmacokinetic evaluation is one of the key processes in drug discovery and development. However, current absorption, distribution, metabolism, excretion prediction models still have limited accuracy. Aim: This study aims to construct an integrated transfer learning and multitask learning approach for developing quantitative structure-activity relationship models to predict four human pharmacokinetic parameters. Methods: A pharmacokinetic dataset included 1104 U.S. FDA approved small molecule drugs. The dataset included four human pharmacokinetic parameter subsets (oral bioavailability, plasma protein binding rate, apparent volume of distribution at steady-state and elimination half-life). The pre-trained model was trained on over 30 million bioactivity data. An integrated transfer learning and multitask learning approach was established to enhance the model generalization. Results: The pharmacokinetic dataset was split into three parts (60:20:20) for training, validation and test by the improved Maximum Dissimilarity algorithm with the representative initial set selection algorithm and the weighted distance function. The multitask learning techniques enhanced the model predictive ability. The integrated transfer learning and multitask learning model demonstrated the best accuracies, because deep neural networks have the general feature extraction ability, transfer learning and multitask learning improved the model generalization. Conclusions: The integrated transfer learning and multitask learning approach with the improved dataset splitting algorithm was firstly introduced to predict the pharmacokinetic parameters. This method can be further employed in drug discovery and development.
Deep learning for in vitro prediction of pharmaceutical formulations
Sep 06, 2018

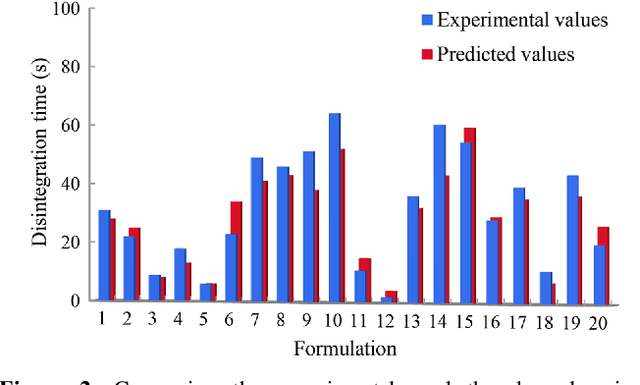
Abstract:Current pharmaceutical formulation development still strongly relies on the traditional trial-and-error approach by individual experiences of pharmaceutical scientists, which is laborious, time-consuming and costly. Recently, deep learning has been widely applied in many challenging domains because of its important capability of automatic feature extraction. The aim of this research is to use deep learning to predict pharmaceutical formulations. In this paper, two different types of dosage forms were chosen as model systems. Evaluation criteria suitable for pharmaceutics were applied to assessing the performance of the models. Moreover, an automatic dataset selection algorithm was developed for selecting the representative data as validation and test datasets. Six machine learning methods were compared with deep learning. The result shows the accuracies of both two deep neural networks were above 80% and higher than other machine learning models, which showed good prediction in pharmaceutical formulations. In summary, deep learning with the automatic data splitting algorithm and the evaluation criteria suitable for pharmaceutical formulation data was firstly developed for the prediction of pharmaceutical formulations. The cross-disciplinary integration of pharmaceutics and artificial intelligence may shift the paradigm of pharmaceutical researches from experience-dependent studies to data-driven methodologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge