Srinandan Dasmahapatra
JetLOV: Enhancing Jet Tree Tagging through Neural Network Learning of Optimal LundNet Variables
Nov 24, 2023Abstract:Machine learning has played a pivotal role in advancing physics, with deep learning notably contributing to solving complex classification problems such as jet tagging in the field of jet physics. In this experiment, we aim to harness the full potential of neural networks while acknowledging that, at times, we may lose sight of the underlying physics governing these models. Nevertheless, we demonstrate that we can achieve remarkable results obscuring physics knowledge and relying completely on the model's outcome. We introduce JetLOV, a composite comprising two models: a straightforward multilayer perceptron (MLP) and the well-established LundNet. Our study reveals that we can attain comparable jet tagging performance without relying on the pre-computed LundNet variables. Instead, we allow the network to autonomously learn an entirely new set of variables, devoid of a priori knowledge of the underlying physics. These findings hold promise, particularly in addressing the issue of model dependence, which can be mitigated through generalization and training on diverse data sets.
Probabilistic Weight Fixing: Large-scale training of neural network weight uncertainties for quantization
Sep 26, 2023Abstract:Weight-sharing quantization has emerged as a technique to reduce energy expenditure during inference in large neural networks by constraining their weights to a limited set of values. However, existing methods for weight-sharing quantization often make assumptions about the treatment of weights based on value alone that neglect the unique role weight position plays. This paper proposes a probabilistic framework based on Bayesian neural networks (BNNs) and a variational relaxation to identify which weights can be moved to which cluster centre and to what degree based on their individual position-specific learned uncertainty distributions. We introduce a new initialisation setting and a regularisation term which allow for the training of BNNs under complex dataset-model combinations. By leveraging the flexibility of weight values captured through a probability distribution, we enhance noise resilience and downstream compressibility. Our iterative clustering procedure demonstrates superior compressibility and higher accuracy compared to state-of-the-art methods on both ResNet models and the more complex transformer-based architectures. In particular, our method outperforms the state-of-the-art quantization method top-1 accuracy by 1.6% on ImageNet using DeiT-Tiny, with its 5 million+ weights now represented by only 296 unique values.
MatSpectNet: Material Segmentation Network with Domain-Aware and Physically-Constrained Hyperspectral Reconstruction
Aug 06, 2023Abstract:Achieving accurate material segmentation for 3-channel RGB images is challenging due to the considerable variation in a material's appearance. Hyperspectral images, which are sets of spectral measurements sampled at multiple wavelengths, theoretically offer distinct information for material identification, as variations in intensity of electromagnetic radiation reflected by a surface depend on the material composition of a scene. However, existing hyperspectral datasets are impoverished regarding the number of images and material categories for the dense material segmentation task, and collecting and annotating hyperspectral images with a spectral camera is prohibitively expensive. To address this, we propose a new model, the MatSpectNet to segment materials with recovered hyperspectral images from RGB images. The network leverages the principles of colour perception in modern cameras to constrain the reconstructed hyperspectral images and employs the domain adaptation method to generalise the hyperspectral reconstruction capability from a spectral recovery dataset to material segmentation datasets. The reconstructed hyperspectral images are further filtered using learned response curves and enhanced with human perception. The performance of MatSpectNet is evaluated on the LMD dataset as well as the OpenSurfaces dataset. Our experiments demonstrate that MatSpectNet attains a 1.60% increase in average pixel accuracy and a 3.42% improvement in mean class accuracy compared with the most recent publication. The project code is attached to the supplementary material and will be published on GitHub.
DBAT: Dynamic Backward Attention Transformer for Material Segmentation with Cross-Resolution Patches
May 06, 2023Abstract:The objective of dense material segmentation is to identify the material categories for every image pixel. Recent studies adopt image patches to extract material features. Although the trained networks can improve the segmentation performance, their methods choose a fixed patch resolution which fails to take into account the variation in pixel area covered by each material. In this paper, we propose the Dynamic Backward Attention Transformer (DBAT) to aggregate cross-resolution features. The DBAT takes cropped image patches as input and gradually increases the patch resolution by merging adjacent patches at each transformer stage, instead of fixing the patch resolution during training. We explicitly gather the intermediate features extracted from cross-resolution patches and merge them dynamically with predicted attention masks. Experiments show that our DBAT achieves an accuracy of 86.85%, which is the best performance among state-of-the-art real-time models. Like other successful deep learning solutions with complex architectures, the DBAT also suffers from lack of interpretability. To address this problem, this paper examines the properties that the DBAT makes use of. By analysing the cross-resolution features and the attention weights, this paper interprets how the DBAT learns from image patches. We further align features to semantic labels, performing network dissection, to infer that the proposed model can extract material-related features better than other methods. We show that the DBAT model is more robust to network initialisation, and yields fewer variable predictions compared to other models. The project code is available at https://github.com/heng-yuwen/Dynamic-Backward-Attention-Transformer.
ADS_UNet: A Nested UNet for Histopathology Image Segmentation
Apr 10, 2023Abstract:The UNet model consists of fully convolutional network (FCN) layers arranged as contracting encoder and upsampling decoder maps. Nested arrangements of these encoder and decoder maps give rise to extensions of the UNet model, such as UNete and UNet++. Other refinements include constraining the outputs of the convolutional layers to discriminate between segment labels when trained end to end, a property called deep supervision. This reduces feature diversity in these nested UNet models despite their large parameter space. Furthermore, for texture segmentation, pixel correlations at multiple scales contribute to the classification task; hence, explicit deep supervision of shallower layers is likely to enhance performance. In this paper, we propose ADS UNet, a stage-wise additive training algorithm that incorporates resource-efficient deep supervision in shallower layers and takes performance-weighted combinations of the sub-UNets to create the segmentation model. We provide empirical evidence on three histopathology datasets to support the claim that the proposed ADS UNet reduces correlations between constituent features and improves performance while being more resource efficient. We demonstrate that ADS_UNet outperforms state-of-the-art Transformer-based models by 1.08 and 0.6 points on CRAG and BCSS datasets, and yet requires only 37% of GPU consumption and 34% of training time as that required by Transformers.
Rotation-Scale Equivariant Steerable Filters
Apr 10, 2023Abstract:Incorporating either rotation equivariance or scale equivariance into CNNs has proved to be effective in improving models' generalization performance. However, jointly integrating rotation and scale equivariance into CNNs has not been widely explored. Digital histology imaging of biopsy tissue can be captured at arbitrary orientation and magnification and stored at different resolutions, resulting in cells appearing in different scales. When conventional CNNs are applied to histopathology image analysis, the generalization performance of models is limited because 1) a part of the parameters of filters are trained to fit rotation transformation, thus decreasing the capability of learning other discriminative features; 2) fixed-size filters trained on images at a given scale fail to generalize to those at different scales. To deal with these issues, we propose the Rotation-Scale Equivariant Steerable Filter (RSESF), which incorporates steerable filters and scale-space theory. The RSESF contains copies of filters that are linear combinations of Gaussian filters, whose direction is controlled by directional derivatives and whose scale parameters are trainable but constrained to span disjoint scales in successive layers of the network. Extensive experiments on two gland segmentation datasets demonstrate that our method outperforms other approaches, with much fewer trainable parameters and fewer GPU resources required. The source code is available at: https://github.com/ynulonger/RSESF.
Scale-Equivariant UNet for Histopathology Image Segmentation
Apr 10, 2023Abstract:Digital histopathology slides are scanned and viewed under different magnifications and stored as images at different resolutions. Convolutional Neural Networks (CNNs) trained on such images at a given scale fail to generalise to those at different scales. This inability is often addressed by augmenting training data with re-scaled images, allowing a model with sufficient capacity to learn the requisite patterns. Alternatively, designing CNN filters to be scale-equivariant frees up model capacity to learn discriminative features. In this paper, we propose the Scale-Equivariant UNet (SEUNet) for image segmentation by building on scale-space theory. The SEUNet contains groups of filters that are linear combinations of Gaussian basis filters, whose scale parameters are trainable but constrained to span disjoint scales through the layers of the network. Extensive experiments on a nuclei segmentation dataset and a tissue type segmentation dataset demonstrate that our method outperforms other approaches, with much fewer trainable parameters.
Weight Fixing Networks
Oct 24, 2022



Abstract:Modern iterations of deep learning models contain millions (billions) of unique parameters, each represented by a b-bit number. Popular attempts at compressing neural networks (such as pruning and quantisation) have shown that many of the parameters are superfluous, which we can remove (pruning) or express with less than b-bits (quantisation) without hindering performance. Here we look to go much further in minimising the information content of networks. Rather than a channel or layer-wise encoding, we look to lossless whole-network quantisation to minimise the entropy and number of unique parameters in a network. We propose a new method, which we call Weight Fixing Networks (WFN) that we design to realise four model outcome objectives: i) very few unique weights, ii) low-entropy weight encodings, iii) unique weight values which are amenable to energy-saving versions of hardware multiplication, and iv) lossless task-performance. Some of these goals are conflicting. To best balance these conflicts, we combine a few novel (and some well-trodden) tricks; a novel regularisation term, (i, ii) a view of clustering cost as relative distance change (i, ii, iv), and a focus on whole-network re-use of weights (i, iii). Our Imagenet experiments demonstrate lossless compression using 56x fewer unique weights and a 1.9x lower weight-space entropy than SOTA quantisation approaches.
Optimising 2D Pose Representation: Improve Accuracy, Stability and Generalisability Within Unsupervised 2D-3D Human Pose Estimation
Sep 01, 2022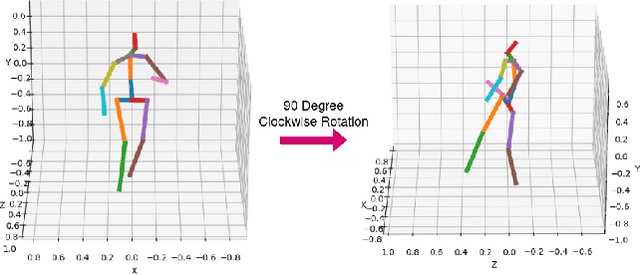
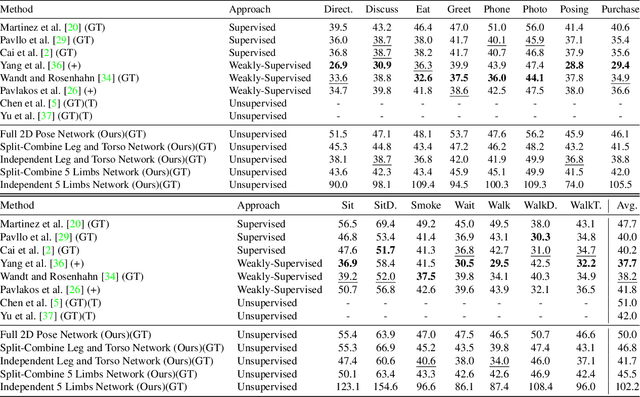

Abstract:This paper addresses the problem of 2D pose representation during unsupervised 2D to 3D pose lifting to improve the accuracy, stability and generalisability of 3D human pose estimation (HPE) models. All unsupervised 2D-3D HPE approaches provide the entire 2D kinematic skeleton to a model during training. We argue that this is sub-optimal and disruptive as long-range correlations are induced between independent 2D key points and predicted 3D ordinates during training. To this end, we conduct the following study. With a maximum architecture capacity of 6 residual blocks, we evaluate the performance of 5 models which each represent a 2D pose differently during the adversarial unsupervised 2D-3D HPE process. Additionally, we show the correlations between 2D key points which are learned during the training process, highlighting the unintuitive correlations induced when an entire 2D pose is provided to a lifting model. Our results show that the most optimal representation of a 2D pose is that of two independent segments, the torso and legs, with no shared features between each lifting network. This approach decreased the average error by 20\% on the Human3.6M dataset when compared to a model with a near identical parameter count trained on the entire 2D kinematic skeleton. Furthermore, due to the complex nature of adversarial learning, we show how this representation can also improve convergence during training allowing for an optimum result to be obtained more often.
"Teaching Independent Parts Separately"(TIPSy-GAN) : Improving Accuracy and Stability in Unsupervised Adversarial 2D to 3D Human Pose Estimation
May 16, 2022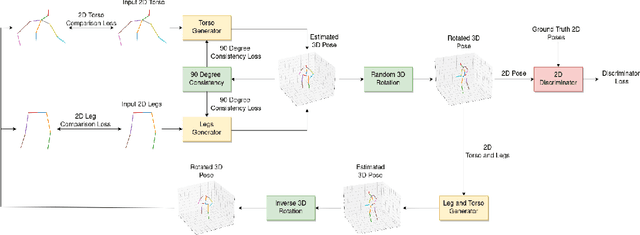
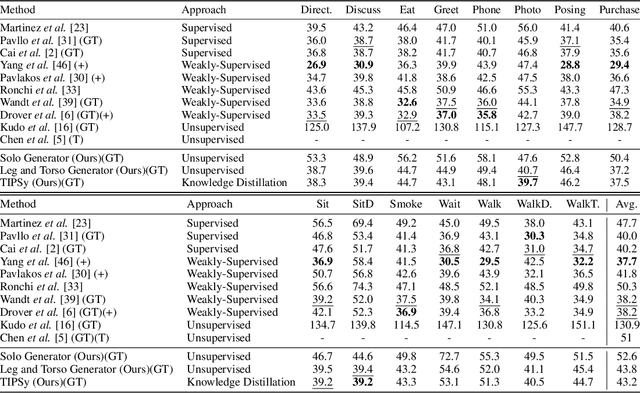
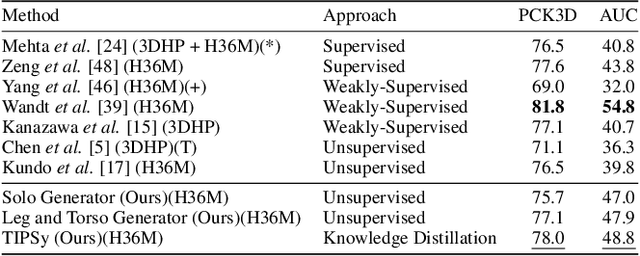
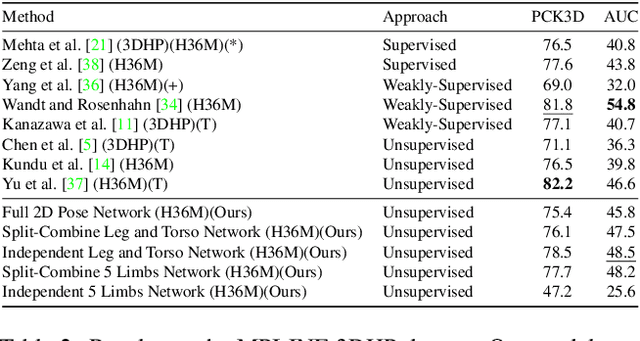
Abstract:We present TIPSy-GAN, a new approach to improve the accuracy and stability in unsupervised adversarial 2D to 3D human pose estimation. In our work we demonstrate that the human kinematic skeleton should not be assumed as one spatially codependent structure. In fact, we believe when a full 2D pose is provided during training, there is an inherent bias learned where the 3D coordinate of a keypoint is spatially codependent on the 2D locations of all other keypoints. To investigate our theory we follow previous adversarial approaches but train two generators on spatially independent parts of the kinematic skeleton, the torso and the legs. We find that improving the 2D reprojection self-consistency cycle is key to lowering the evaluation error and therefore introduce new consistency constraints during training. A TIPSy is produced model via knowledge distillation from these generators which can predict the 3D coordinates for the entire 2D pose with improved results. Furthermore, we address the question left unanswered in prior work detailing how long to train for a truly unsupervised scenario. We show that two independent generators training adversarially has improved stability than that of a solo generator which will collapse due to the adversarial network becoming unstable. TIPSy decreases the average error by 18% when compared to that of a baseline solo generator. TIPSy improves upon other unsupervised approaches while also performing strongly against supervised and weakly-supervised approaches during evaluation on both the Human3.6M and MPI-INF-3DHP dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge