Yiying Wang
Transferable Graph Condensation from the Causal Perspective
Jan 29, 2026Abstract:The increasing scale of graph datasets has significantly improved the performance of graph representation learning methods, but it has also introduced substantial training challenges. Graph dataset condensation techniques have emerged to compress large datasets into smaller yet information-rich datasets, while maintaining similar test performance. However, these methods strictly require downstream applications to match the original dataset and task, which often fails in cross-task and cross-domain scenarios. To address these challenges, we propose a novel causal-invariance-based and transferable graph dataset condensation method, named \textbf{TGCC}, providing effective and transferable condensed datasets. Specifically, to preserve domain-invariant knowledge, we first extract domain causal-invariant features from the spatial domain of the graph using causal interventions. Then, to fully capture the structural and feature information of the original graph, we perform enhanced condensation operations. Finally, through spectral-domain enhanced contrastive learning, we inject the causal-invariant features into the condensed graph, ensuring that the compressed graph retains the causal information of the original graph. Experimental results on five public datasets and our novel \textbf{FinReport} dataset demonstrate that TGCC achieves up to a 13.41\% improvement in cross-task and cross-domain complex scenarios compared to existing methods, and achieves state-of-the-art performance on 5 out of 6 datasets in the single dataset and task scenario.
NeCA: 3D Coronary Artery Tree Reconstruction from Two 2D Projections by Neural Implicit Representation
Sep 06, 2024Abstract:Cardiovascular diseases (CVDs) are the most common health threats worldwide. 2D x-ray invasive coronary angiography (ICA) remains as the most widely adopted imaging modality for CVDs diagnosis. However, in current clinical practice, it is often difficult for the cardiologists to interpret the 3D geometry of coronary vessels based on 2D planes. Moreover, due to the radiation limit, in general only two angiographic projections are acquired, providing limited information of the vessel geometry and necessitating 3D coronary tree reconstruction based only on two ICA projections. In this paper, we propose a self-supervised deep learning method called NeCA, which is based on implicit neural representation using the multiresolution hash encoder and differentiable cone-beam forward projector layer in order to achieve 3D coronary artery tree reconstruction from two projections. We validate our method using six different metrics on coronary computed tomography angiography data in terms of right coronary artery and left anterior descending respectively. The evaluation results demonstrate that our NeCA method, without 3D ground truth for supervision and large datasets for training, achieves promising performance in both vessel topology preservation and branch-connectivity maintaining compared to the supervised deep learning model.
Deep Learning-based 3D Coronary Tree Reconstruction from Two 2D Non-simultaneous X-ray Angiography Projections
Jul 19, 2024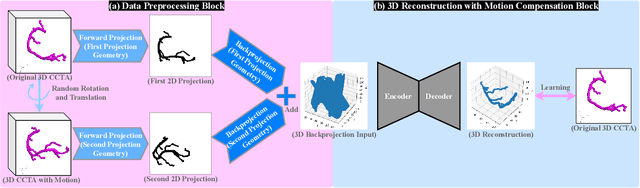

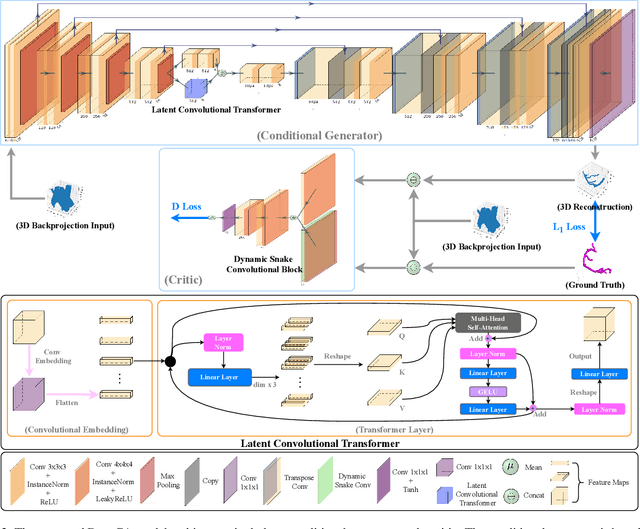

Abstract:Cardiovascular diseases (CVDs) are the most common cause of death worldwide. Invasive x-ray coronary angiography (ICA) is one of the most important imaging modalities for the diagnosis of CVDs. ICA typically acquires only two 2D projections, which makes the 3D geometry of coronary vessels difficult to interpret, thus requiring 3D coronary tree reconstruction from two projections. State-of-the-art approaches require significant manual interactions and cannot correct the non-rigid cardiac and respiratory motions between non-simultaneous projections. In this study, we propose a novel deep learning pipeline. We leverage the Wasserstein conditional generative adversarial network with gradient penalty, latent convolutional transformer layers, and a dynamic snake convolutional critic to implicitly compensate for the non-rigid motion and provide 3D coronary tree reconstruction. Through simulating projections from coronary computed tomography angiography (CCTA), we achieve the generalisation of 3D coronary tree reconstruction on real non-simultaneous ICA projections. We incorporate an application-specific evaluation metric to validate our proposed model on both a CCTA dataset and a real ICA dataset, together with Chamfer L1 distance. The results demonstrate the good performance of our model in vessel topology preservation, recovery of missing features, and generalisation ability to real ICA data. To the best of our knowledge, this is the first study that leverages deep learning to achieve 3D coronary tree reconstruction from two real non-simultaneous x-ray angiography projections.
PEER: Expertizing Domain-Specific Tasks with a Multi-Agent Framework and Tuning Methods
Jul 10, 2024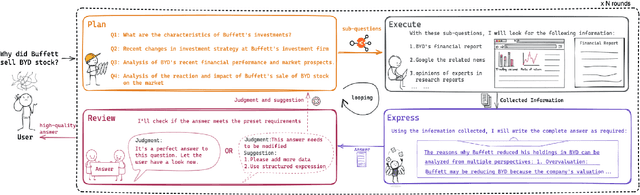

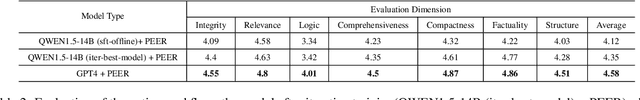
Abstract:In domain-specific applications, GPT-4, augmented with precise prompts or Retrieval-Augmented Generation (RAG), shows notable potential but faces the critical tri-lemma of performance, cost, and data privacy. High performance requires sophisticated processing techniques, yet managing multiple agents within a complex workflow often proves costly and challenging. To address this, we introduce the PEER (Plan, Execute, Express, Review) multi-agent framework. This systematizes domain-specific tasks by integrating precise question decomposition, advanced information retrieval, comprehensive summarization, and rigorous self-assessment. Given the concerns of cost and data privacy, enterprises are shifting from proprietary models like GPT-4 to custom models, striking a balance between cost, security, and performance. We developed industrial practices leveraging online data and user feedback for efficient model tuning. This study provides best practice guidelines for applying multi-agent systems in domain-specific problem-solving and implementing effective agent tuning strategies. Our empirical studies, particularly in the financial question-answering domain, demonstrate that our approach achieves 95.0% of GPT-4's performance, while effectively managing costs and ensuring data privacy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge