Sina Tabakhi
Interpretable Multimodal Learning for Tumor Protein-Metal Binding: Progress, Challenges, and Perspectives
Apr 04, 2025Abstract:In cancer therapeutics, protein-metal binding mechanisms critically govern drug pharmacokinetics and targeting efficacy, thereby fundamentally shaping the rational design of anticancer metallodrugs. While conventional laboratory methods used to study such mechanisms are often costly, low throughput, and limited in capturing dynamic biological processes, machine learning (ML) has emerged as a promising alternative. Despite increasing efforts to develop protein-metal binding datasets and ML algorithms, the application of ML in tumor protein-metal binding remains limited. Key challenges include a shortage of high-quality, tumor-specific datasets, insufficient consideration of multiple data modalities, and the complexity of interpreting results due to the ''black box'' nature of complex ML models. This paper summarizes recent progress and ongoing challenges in using ML to predict tumor protein-metal binding, focusing on data, modeling, and interpretability. We present multimodal protein-metal binding datasets and outline strategies for acquiring, curating, and preprocessing them for training ML models. Moreover, we explore the complementary value provided by different data modalities and examine methods for their integration. We also review approaches for improving model interpretability to support more trustworthy decisions in cancer research. Finally, we offer our perspective on research opportunities and propose strategies to address the scarcity of tumor protein data and the limited number of predictive models for tumor protein-metal binding. We also highlight two promising directions for effective metal-based drug design: integrating protein-protein interaction data to provide structural insights into metal-binding events and predicting structural changes in tumor proteins after metal binding.
Towards deployment-centric multimodal AI beyond vision and language
Apr 04, 2025Abstract:Multimodal artificial intelligence (AI) integrates diverse types of data via machine learning to improve understanding, prediction, and decision-making across disciplines such as healthcare, science, and engineering. However, most multimodal AI advances focus on models for vision and language data, while their deployability remains a key challenge. We advocate a deployment-centric workflow that incorporates deployment constraints early to reduce the likelihood of undeployable solutions, complementing data-centric and model-centric approaches. We also emphasise deeper integration across multiple levels of multimodality and multidisciplinary collaboration to significantly broaden the research scope beyond vision and language. To facilitate this approach, we identify common multimodal-AI-specific challenges shared across disciplines and examine three real-world use cases: pandemic response, self-driving car design, and climate change adaptation, drawing expertise from healthcare, social science, engineering, science, sustainability, and finance. By fostering multidisciplinary dialogue and open research practices, our community can accelerate deployment-centric development for broad societal impact.
Co-evolution-based Metal-binding Residue Prediction with Graph Neural Networks
Feb 22, 2025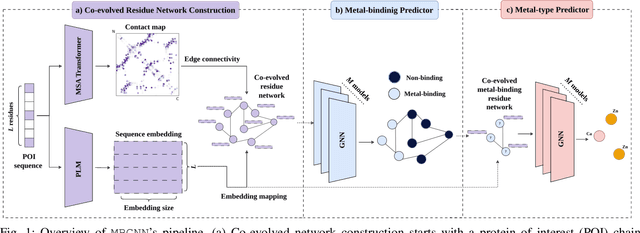
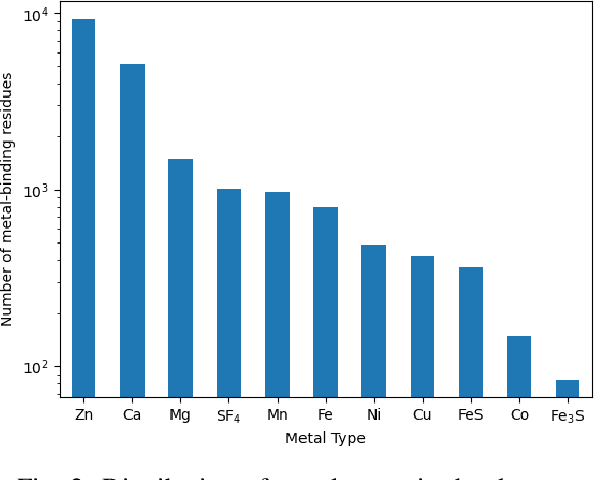
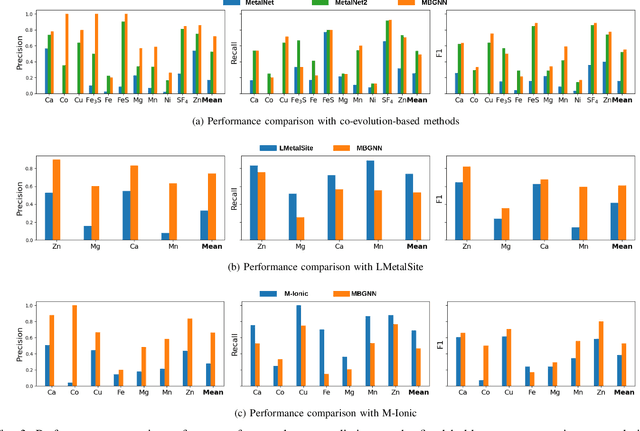
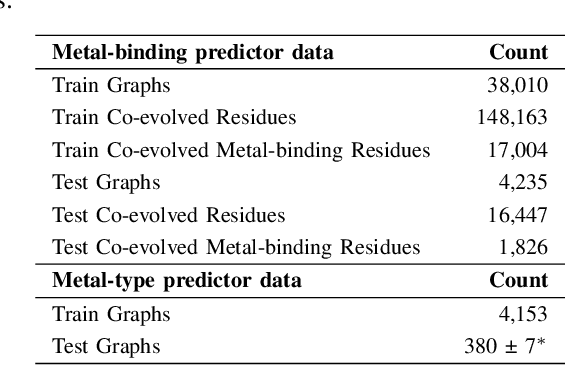
Abstract:In computational structural biology, predicting metal-binding sites and their corresponding metal types is challenging due to the complexity of protein structures and interactions. Conventional sequence- and structure-based prediction approaches cannot capture the complex evolutionary relationships driving these interactions to facilitate understanding, while recent co-evolution-based approaches do not fully consider the entire structure of the co-evolved residue network. In this paper, we introduce MBGNN (Metal-Binding Graph Neural Network) that utilizes the entire co-evolved residue network and effectively captures the complex dependencies within protein structures via graph neural networks to enhance the prediction of co-evolved metal-binding residues and their associated metal types. Experimental results on a public dataset show that MBGNN outperforms existing co-evolution-based metal-binding prediction methods, and it is also competitive against recent sequence-based methods, showing the potential of integrating co-evolutionary insights with advanced machine learning to deepen our understanding of protein-metal interactions. The MBGNN code is publicly available at https://github.com/SRastegari/MBGNN.
Classifying the Stoichiometry of Virus-like Particles with Interpretable Machine Learning
Feb 17, 2025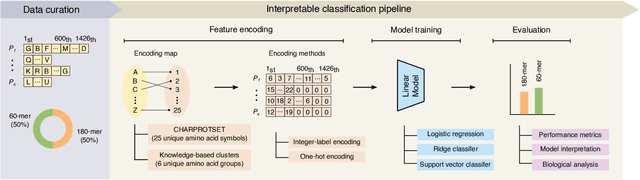
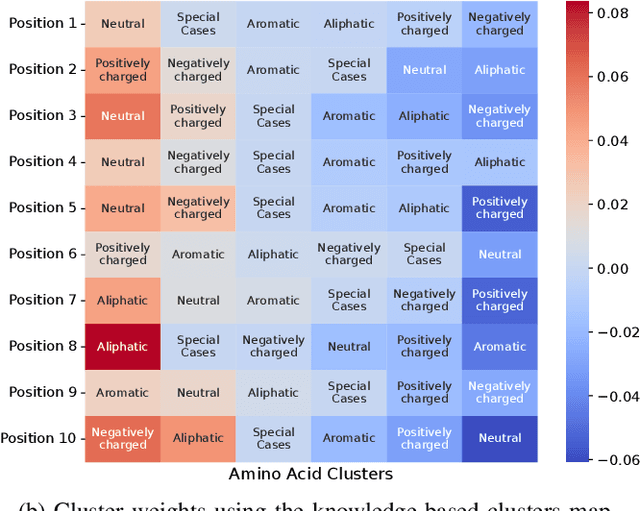
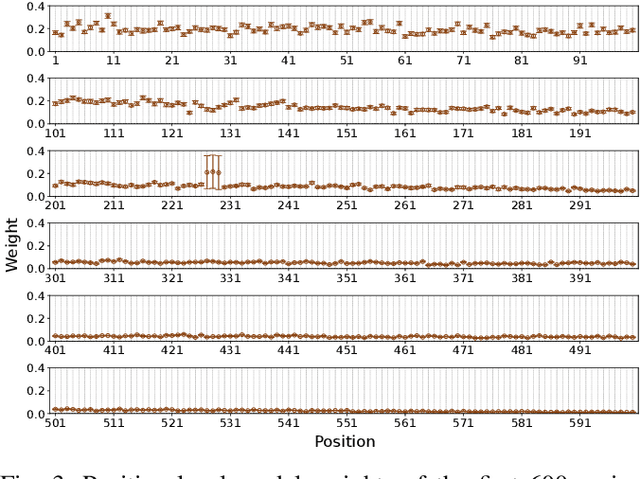
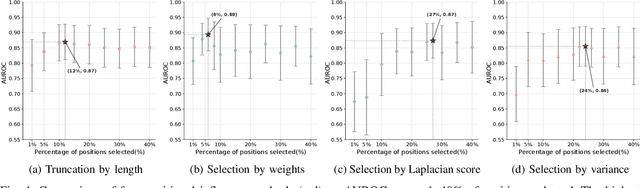
Abstract:Virus-like particles (VLPs) are valuable for vaccine development due to their immune-triggering properties. Understanding their stoichiometry, the number of protein subunits to form a VLP, is critical for vaccine optimisation. However, current experimental methods to determine stoichiometry are time-consuming and require highly purified proteins. To efficiently classify stoichiometry classes in proteins, we curate a new dataset and propose an interpretable, data-driven pipeline leveraging linear machine learning models. We also explore the impact of feature encoding on model performance and interpretability, as well as methods to identify key protein sequence features influencing classification. The evaluation of our pipeline demonstrates that it can classify stoichiometry while revealing protein features that possibly influence VLP assembly. The data and code used in this work are publicly available at https://github.com/Shef-AIRE/StoicIML.
Heterogeneous graph attention network improves cancer multiomics integration
Aug 05, 2024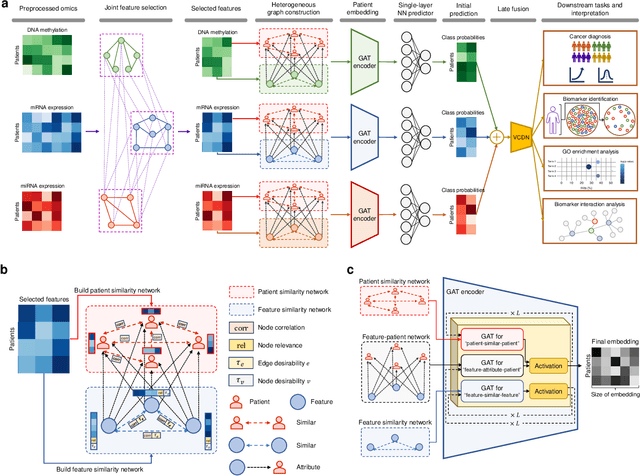
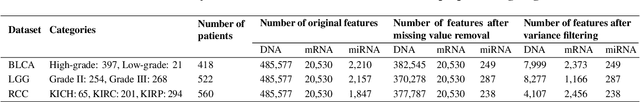
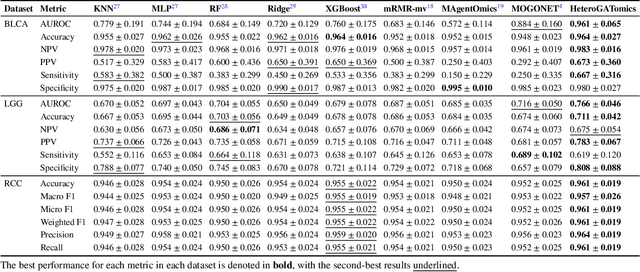
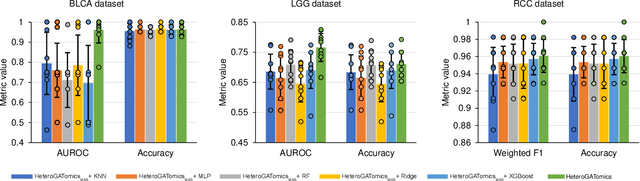
Abstract:The increase in high-dimensional multiomics data demands advanced integration models to capture the complexity of human diseases. Graph-based deep learning integration models, despite their promise, struggle with small patient cohorts and high-dimensional features, often applying independent feature selection without modeling relationships among omics. Furthermore, conventional graph-based omics models focus on homogeneous graphs, lacking multiple types of nodes and edges to capture diverse structures. We introduce a Heterogeneous Graph ATtention network for omics integration (HeteroGATomics) to improve cancer diagnosis. HeteroGATomics performs joint feature selection through a multi-agent system, creating dedicated networks of feature and patient similarity for each omic modality. These networks are then combined into one heterogeneous graph for learning holistic omic-specific representations and integrating predictions across modalities. Experiments on three cancer multiomics datasets demonstrate HeteroGATomics' superior performance in cancer diagnosis. Moreover, HeteroGATomics enhances interpretability by identifying important biomarkers contributing to the diagnosis outcomes.
Interpretable Multimodal Learning for Cardiovascular Hemodynamics Assessment
Apr 06, 2024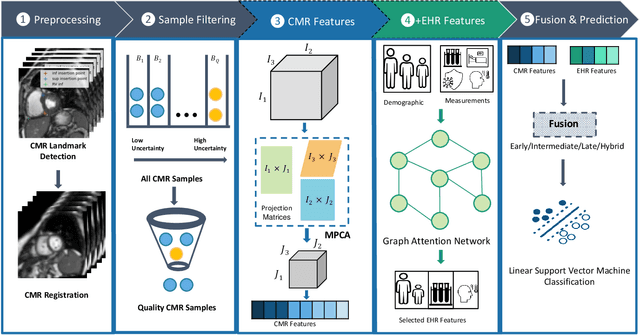
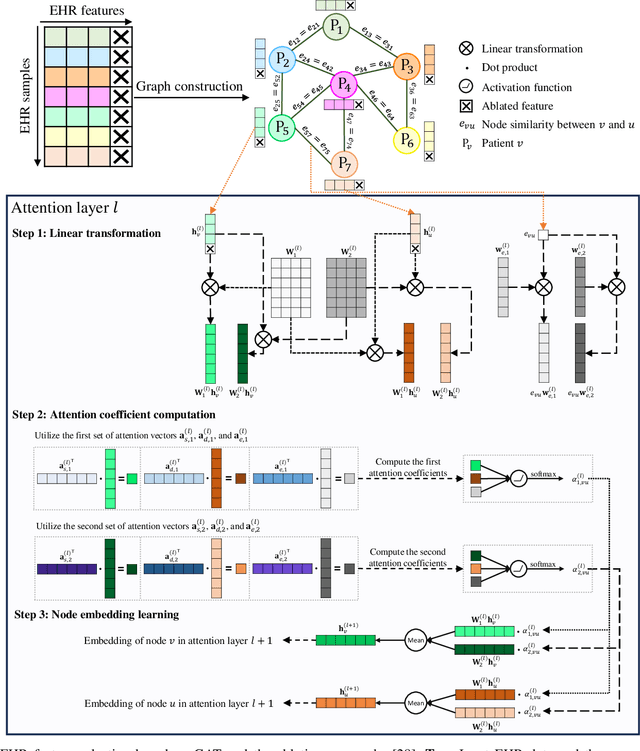
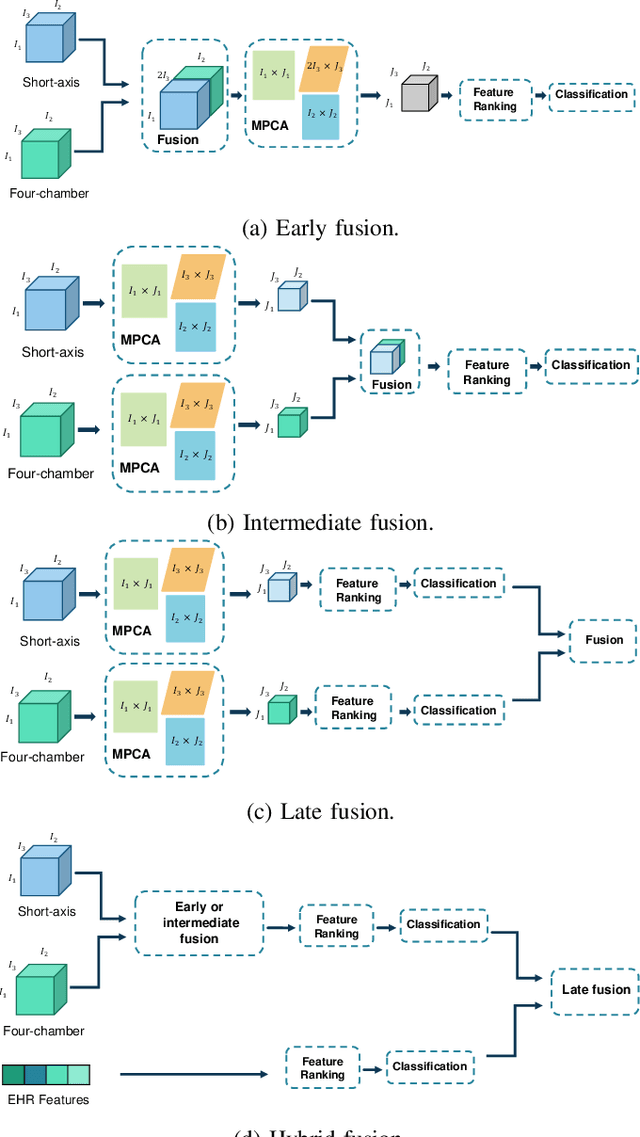
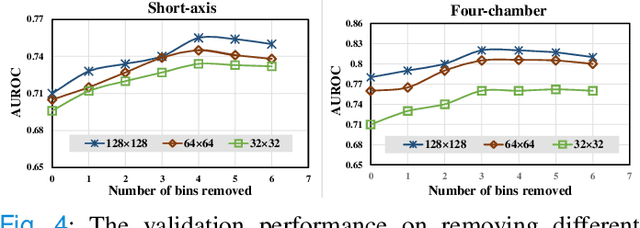
Abstract:Pulmonary Arterial Wedge Pressure (PAWP) is an essential cardiovascular hemodynamics marker to detect heart failure. In clinical practice, Right Heart Catheterization is considered a gold standard for assessing cardiac hemodynamics while non-invasive methods are often needed to screen high-risk patients from a large population. In this paper, we propose a multimodal learning pipeline to predict PAWP marker. We utilize complementary information from Cardiac Magnetic Resonance Imaging (CMR) scans (short-axis and four-chamber) and Electronic Health Records (EHRs). We extract spatio-temporal features from CMR scans using tensor-based learning. We propose a graph attention network to select important EHR features for prediction, where we model subjects as graph nodes and feature relationships as graph edges using the attention mechanism. We design four feature fusion strategies: early, intermediate, late, and hybrid fusion. With a linear classifier and linear fusion strategies, our pipeline is interpretable. We validate our pipeline on a large dataset of $2,641$ subjects from our ASPIRE registry. The comparative study against state-of-the-art methods confirms the superiority of our pipeline. The decision curve analysis further validates that our pipeline can be applied to screen a large population. The code is available at https://github.com/prasunc/hemodynamics.
Multimodal Learning for Multi-Omics: A Survey
Dec 19, 2022Abstract:With advanced imaging, sequencing, and profiling technologies, multiple omics data become increasingly available and hold promises for many healthcare applications such as cancer diagnosis and treatment. Multimodal learning for integrative multi-omics analysis can help researchers and practitioners gain deep insights into human diseases and improve clinical decisions. However, several challenges are hindering the development in this area, including the availability of easily accessible open-source tools. This survey aims to provide an up-to-date overview of the data challenges, fusion approaches, datasets, and software tools from several new perspectives. We identify and investigate various omics data challenges that can help us understand the field better. We categorize fusion approaches comprehensively to cover existing methods in this area. We collect existing open-source tools to facilitate their broader utilization and development. We explore a broad range of omics data modalities and a list of accessible datasets. Finally, we summarize future directions that can potentially address existing gaps and answer the pressing need to advance multimodal learning for multi-omics data analysis.
Universal Feature Selection Tool (UniFeat): An Open-Source Tool for Dimensionality Reduction
Nov 30, 2022Abstract:The Universal Feature Selection Tool (UniFeat) is an open-source tool developed entirely in Java for performing feature selection processes in various research areas. It provides a set of well-known and advanced feature selection methods within its significant auxiliary tools. This allows users to compare the performance of feature selection methods. Moreover, due to the open-source nature of UniFeat, researchers can use and modify it in their research, which facilitates the rapid development of new feature selection algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge