David Weston
Incorporating Dictionaries into a Neural Network Architecture to Extract COVID-19 Medical Concepts From Social Media
Sep 05, 2023Abstract:We investigate the potential benefit of incorporating dictionary information into a neural network architecture for natural language processing. In particular, we make use of this architecture to extract several concepts related to COVID-19 from an on-line medical forum. We use a sample from the forum to manually curate one dictionary for each concept. In addition, we use MetaMap, which is a tool for extracting biomedical concepts, to identify a small number of semantic concepts. For a supervised concept extraction task on the forum data, our best model achieved a macro $F_1$ score of 90\%. A major difficulty in medical concept extraction is obtaining labelled data from which to build supervised models. We investigate the utility of our models to transfer to data derived from a different source in two ways. First for producing labels via weak learning and second to perform concept extraction. The dataset we use in this case comprises COVID-19 related tweets and we achieve an $F_1$ score 81\% for symptom concept extraction trained on weakly labelled data. The utility of our dictionaries is compared with a COVID-19 symptom dictionary that was constructed directly from Twitter. Further experiments that incorporate BERT and a COVID-19 version of BERTweet demonstrate that the dictionaries provide a commensurate result. Our results show that incorporating small domain dictionaries to deep learning models can improve concept extraction tasks. Moreover, models built using dictionaries generalize well and are transferable to different datasets on a similar task.
Triage and diagnosis of COVID-19 from medical social media
Mar 22, 2021
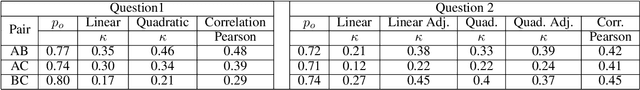
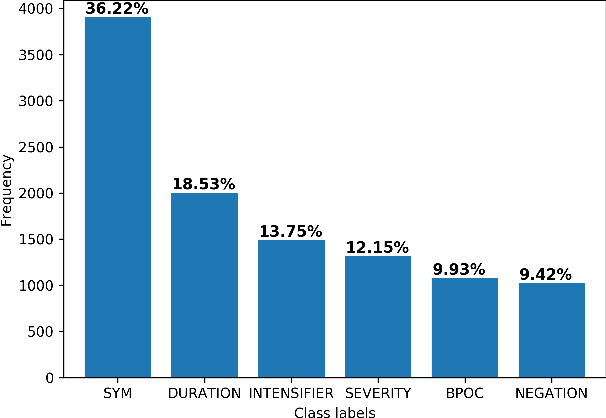
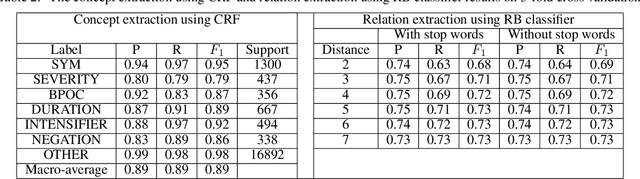
Abstract:Objective: This study aims to develop an end-to-end natural language processing pipeline for triage and diagnosis of COVID-19 from patient-authored social media posts. Materials and Methods: The text processing pipeline first extracts COVID-19 symptoms and related concepts such as severity, duration, negations, and body parts from patients posts using conditional random fields. An unsupervised rule-based algorithm is then applied to establish relations between concepts in the next step of the pipeline. The extracted concepts and relations are subsequently used to construct two different vector representations of each post. These vectors are applied separately to build support vector machine learning models to triage patients into three categories and diagnose them for COVID-19. Results: We report that Macro- and Micro-averaged F_1 scores in the range of 71-96% and 61-87%, respectively, for the triage and diagnosis of COVID-19, when the models are trained on ground truth labelled data. Our experimental results indicate that similar performance can be achieved when the models are trained using predicted labels from concept extraction and rule-based classifiers, thus yielding end-to-end machine learning. Discussion: We highlight important features uncovered by our diagnostic machine learning models and compare them with the most frequent symptoms revealed in another COVID-19 dataset. In particular, we found that the most important features are not always the most frequent ones. Conclusions: Our preliminary results show that it is possible to automatically triage and diagnose patients for COVID-19 from natural language narratives using a machine learning pipeline.
Supervised Phrase-boundary Embeddings
Feb 15, 2020
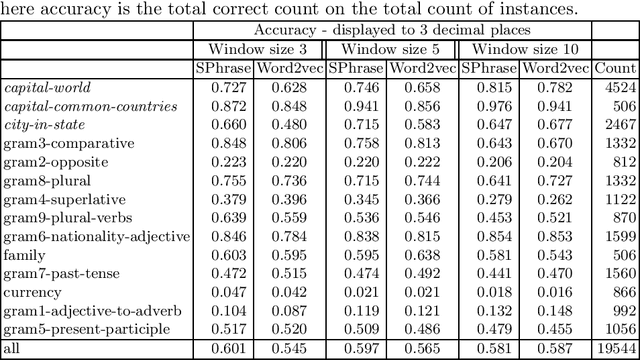

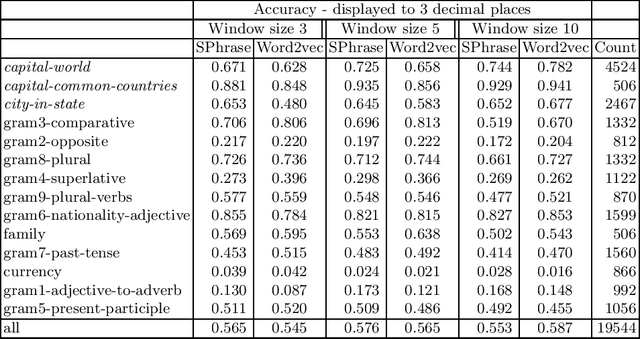
Abstract:We propose a new word embedding model, called SPhrase, that incorporates supervised phrase information. Our method modifies traditional word embeddings by ensuring that all target words in a phrase have exactly the same context. We demonstrate that including this information within a context window produces superior embeddings for both intrinsic evaluation tasks and downstream extrinsic tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge