Md Tauhidul Islam
Deep-and-Wide Learning: Enhancing Data-Driven Inference via Synergistic Learning of Inter- and Intra-Data Representations
Jan 28, 2025Abstract:Advancements in deep learning are revolutionizing science and engineering. The immense success of deep learning is largely due to its ability to extract essential high-dimensional (HD) features from input data and make inference decisions based on this information. However, current deep neural network (DNN) models face several challenges, such as the requirements of extensive amounts of data and computational resources. Here, we introduce a new learning scheme, referred to as deep-and-wide learning (DWL), to systematically capture features not only within individual input data (intra-data features) but also across the data (inter-data features). Furthermore, we propose a dual-interactive-channel network (D-Net) to realize the DWL, which leverages our Bayesian formulation of low-dimensional (LD) inter-data feature extraction and its synergistic interaction with the conventional HD representation of the dataset, for substantially enhanced computational efficiency and inference. The proposed technique has been applied to data across various disciplines for both classification and regression tasks. Our results demonstrate that DWL surpasses state-of-the-art DNNs in accuracy by a substantial margin with limited training data and improves the computational efficiency by order(s) of magnitude. The proposed DWL strategy dramatically alters the data-driven learning techniques, including emerging large foundation models, and sheds significant insights into the evolving field of AI.
Discovering distinctive elements of biomedical datasets for high-performance exploration
Oct 07, 2024



Abstract:The human brain represents an object by small elements and distinguishes two objects based on the difference in elements. Discovering the distinctive elements of high-dimensional datasets is therefore critical in numerous perception-driven biomedical and clinical studies. However, currently there is no available method for reliable extraction of distinctive elements of high-dimensional biomedical and clinical datasets. Here we present an unsupervised deep learning technique namely distinctive element analysis (DEA), which extracts the distinctive data elements using high-dimensional correlative information of the datasets. DEA at first computes a large number of distinctive parts of the data, then filters and condenses the parts into DEA elements by employing a unique kernel-driven triple-optimization network. DEA has been found to improve the accuracy by up to 45% in comparison to the traditional techniques in applications such as disease detection from medical images, gene ranking and cell recognition from single cell RNA sequence (scRNA-seq) datasets. Moreover, DEA allows user-guided manipulation of the intermediate calculation process and thus offers intermediate results with better interpretability.
A Hybrid Deep Feature-Based Deformable Image Registration Method for Pathological Images
Aug 17, 2022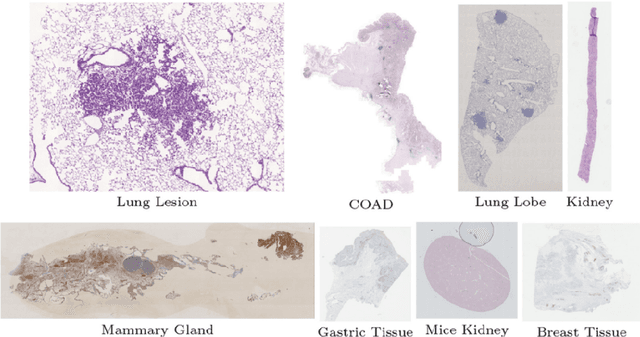
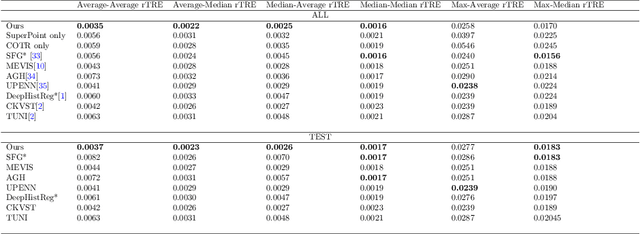
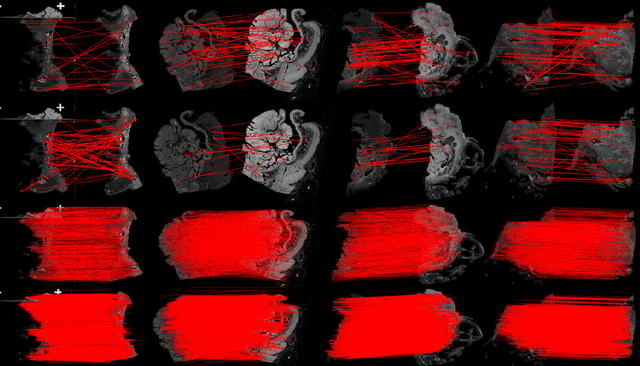
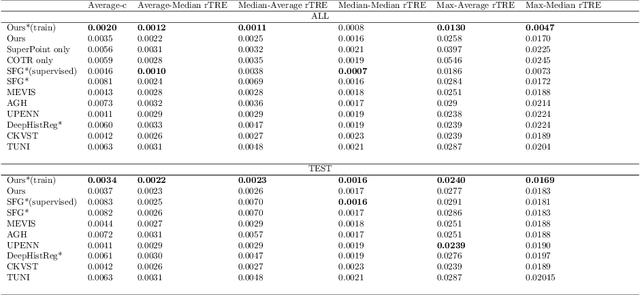
Abstract:Pathologists need to combine information from differently stained pathological slices to obtain accurate diagnostic results. Deformable image registration is a necessary technique for fusing multi-modal pathological slices. This paper proposes a hybrid deep feature-based deformable image registration framework for stained pathological samples. We first extract dense feature points and perform points matching by two deep learning feature networks. Then, to further reduce false matches, an outlier detection method combining the isolation forest statistical model and the local affine correction model is proposed. Finally, the interpolation method generates the DVF for pathology image registration based on the above matching points. We evaluate our method on the dataset of the Non-rigid Histology Image Registration (ANHIR) challenge, which is co-organized with the IEEE ISBI 2019 conference. Our technique outperforms the traditional approaches by 17% with the Average-Average registration target error (rTRE) reaching 0.0034. The proposed method achieved state-of-the-art performance and ranking it 1 in evaluating the test dataset. The proposed hybrid deep feature-based registration method can potentially become a reliable method for pathology image registration.
Learning Treatment Plan Representations for Content Based Image Retrieval
Jun 06, 2022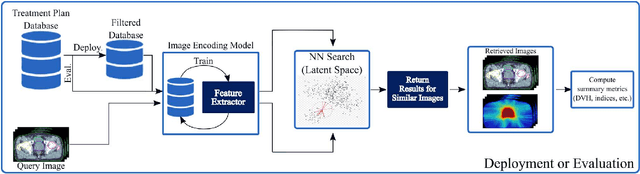
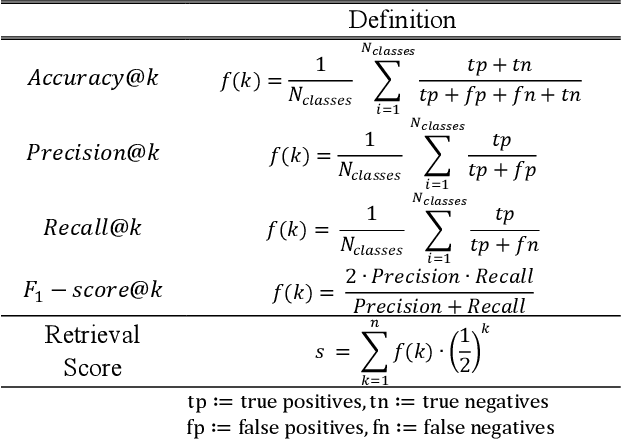
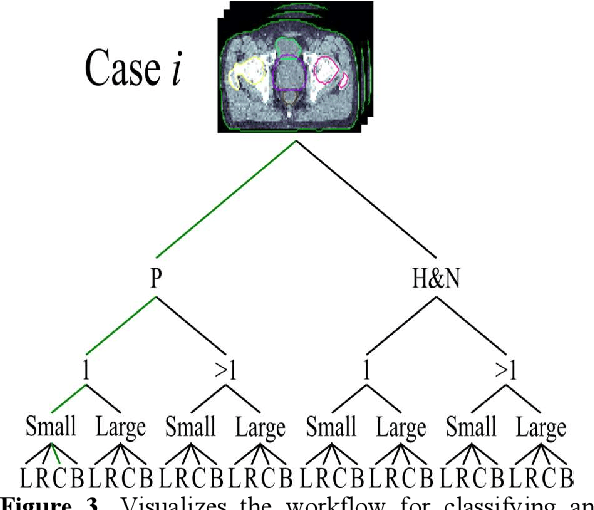
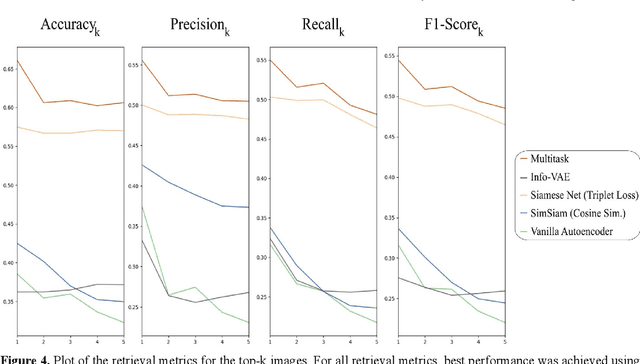
Abstract:Objective: Knowledge based planning (KBP) typically involves training an end-to-end deep learning model to predict dose distributions. However, training end-to-end KBP methods may be associated with practical limitations due to the limited size of medical datasets that are often used. To address these limitations, we propose a content based image retrieval (CBIR) method for retrieving dose distributions of previously planned patients based on anatomical similarity. Approach: Our proposed CBIR method trains a representation model that produces latent space embeddings of a patient's anatomical information. The latent space embeddings of new patients are then compared against those of previous patients in a database for image retrieval of dose distributions. Summary metrics (e.g. dose-volume histogram, conformity index, homogeneity index, etc.) are computed and can then be utilized in subsequent automated planning. All source code for this project is available on github. Main Results: The retrieval performance of various CBIR methods is evaluated on a dataset consisting of both publicly available plans and clinical plans from our institution. This study compares various encoding methods, ranging from simple autoencoders to more recent Siamese networks like SimSiam, and the best performance was observed for the multitask Siamese network. Significance: Applying CBIR to inform subsequent treatment planning potentially addresses many limitations associated with end-to-end KBP. Our current results demonstrate that excellent image retrieval performance can be obtained through slight changes to previously developed Siamese networks. We hope to integrate CBIR into automated planning workflow in future works, potentially through methods like the MetaPlanner framework.
Image Classification using Graph Neural Network and Multiscale Wavelet Superpixels
Jan 29, 2022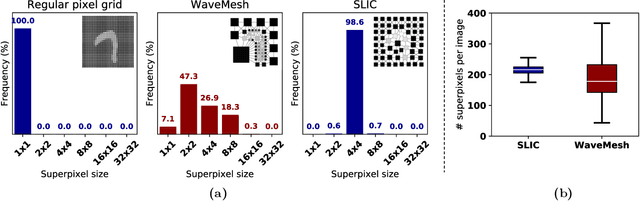
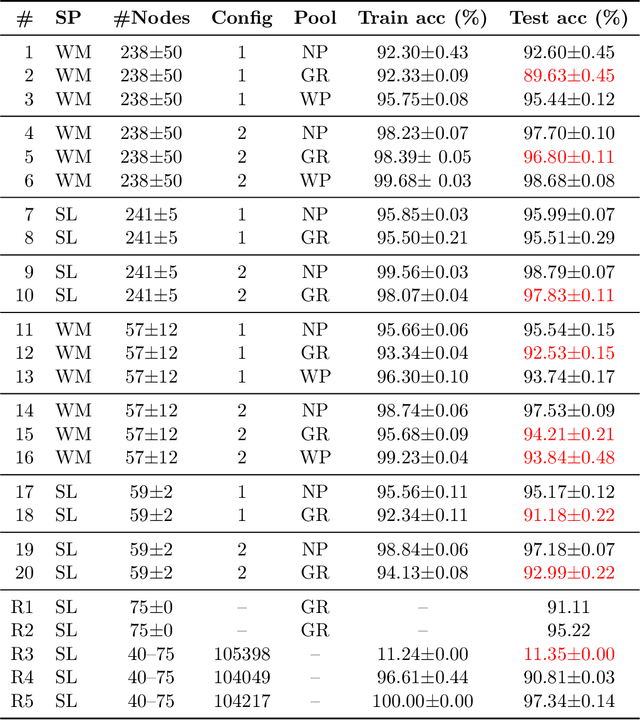
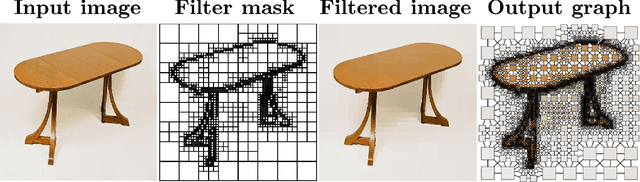
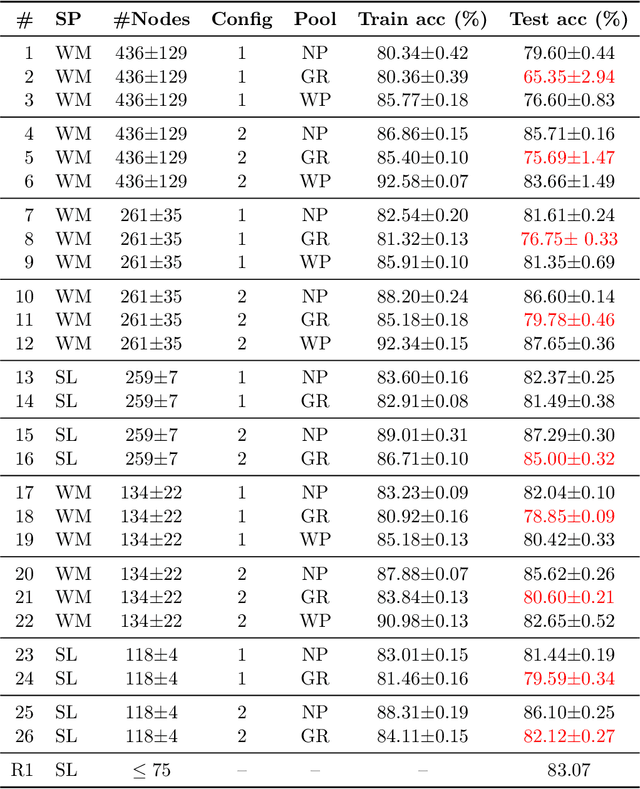
Abstract:Prior studies using graph neural networks (GNNs) for image classification have focused on graphs generated from a regular grid of pixels or similar-sized superpixels. In the latter, a single target number of superpixels is defined for an entire dataset irrespective of differences across images and their intrinsic multiscale structure. On the contrary, this study investigates image classification using graphs generated from an image-specific number of multiscale superpixels. We propose WaveMesh, a new wavelet-based superpixeling algorithm, where the number and sizes of superpixels in an image are systematically computed based on its content. WaveMesh superpixel graphs are structurally different from similar-sized superpixel graphs. We use SplineCNN, a state-of-the-art network for image graph classification, to compare WaveMesh and similar-sized superpixels. Using SplineCNN, we perform extensive experiments on three benchmark datasets under three local-pooling settings: 1) no pooling, 2) GraclusPool, and 3) WavePool, a novel spatially heterogeneous pooling scheme tailored to WaveMesh superpixels. Our experiments demonstrate that SplineCNN learns from multiscale WaveMesh superpixels on-par with similar-sized superpixels. In all WaveMesh experiments, GraclusPool performs poorer than no pooling / WavePool, indicating that poor choice of pooling can result in inferior performance while learning from multiscale superpixels.
Self-supervised Feature Learning via Exploiting Multi-modal Data for Retinal Disease Diagnosis
Jul 21, 2020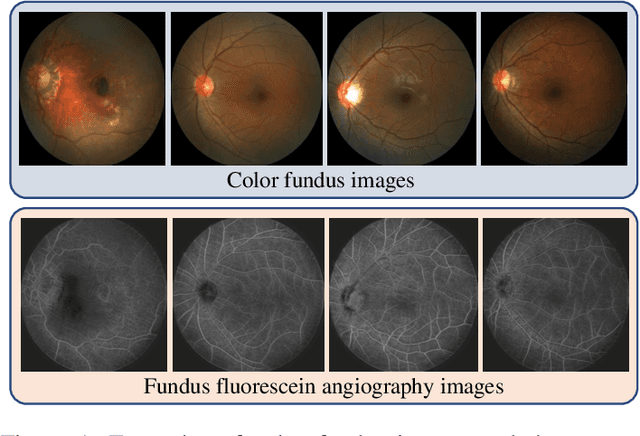
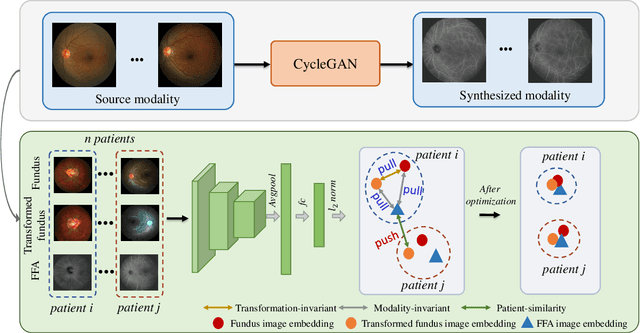
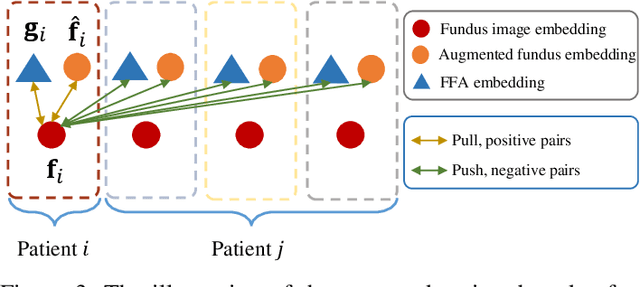
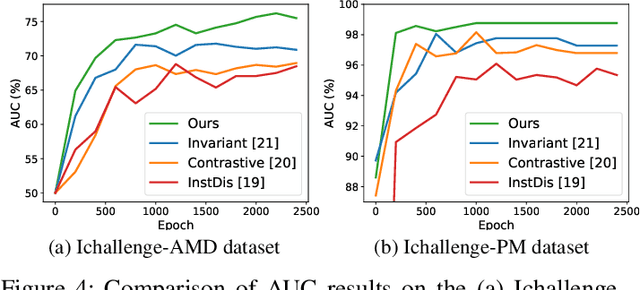
Abstract:The automatic diagnosis of various retinal diseases from fundus images is important to support clinical decision-making. However, developing such automatic solutions is challenging due to the requirement of a large amount of human-annotated data. Recently, unsupervised/self-supervised feature learning techniques receive a lot of attention, as they do not need massive annotations. Most of the current self-supervised methods are analyzed with single imaging modality and there is no method currently utilize multi-modal images for better results. Considering that the diagnostics of various vitreoretinal diseases can greatly benefit from another imaging modality, e.g., FFA, this paper presents a novel self-supervised feature learning method by effectively exploiting multi-modal data for retinal disease diagnosis. To achieve this, we first synthesize the corresponding FFA modality and then formulate a patient feature-based softmax embedding objective. Our objective learns both modality-invariant features and patient-similarity features. Through this mechanism, the neural network captures the semantically shared information across different modalities and the apparent visual similarity between patients. We evaluate our method on two public benchmark datasets for retinal disease diagnosis. The experimental results demonstrate that our method clearly outperforms other self-supervised feature learning methods and is comparable to the supervised baseline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge