Maxime Bassenne
Image Classification using Graph Neural Network and Multiscale Wavelet Superpixels
Jan 29, 2022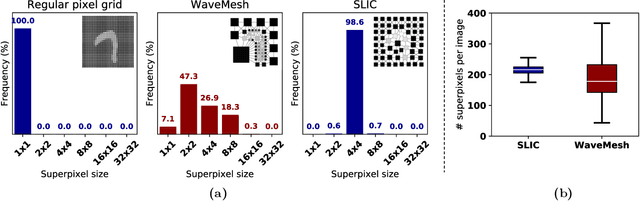
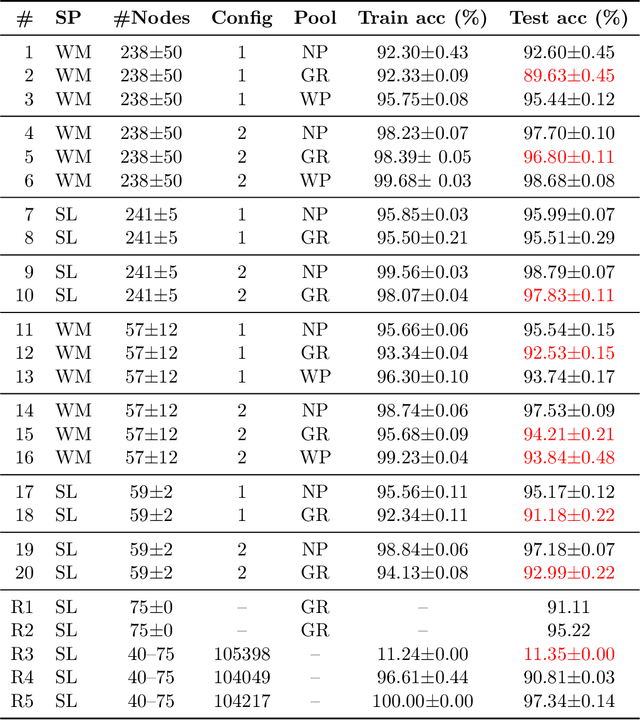
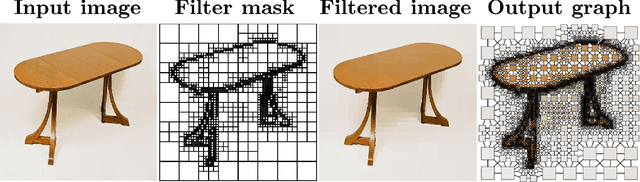
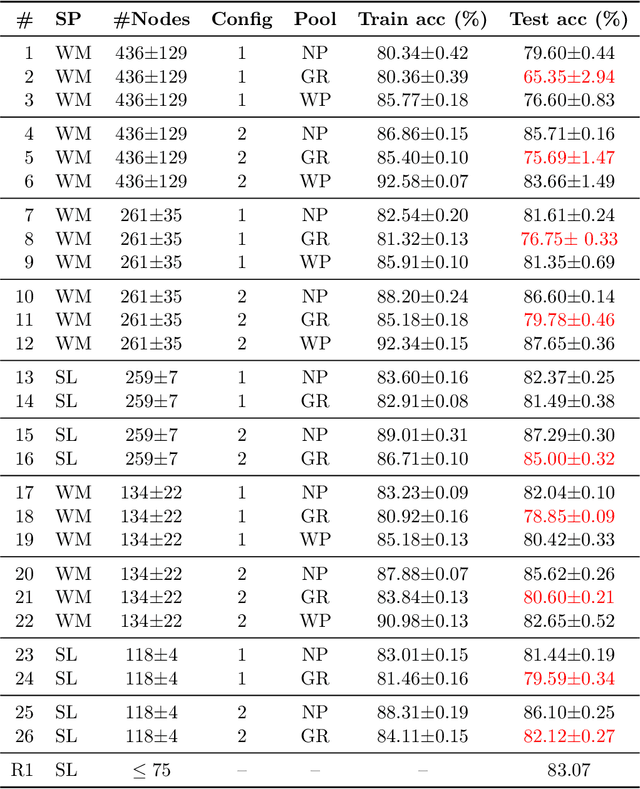
Abstract:Prior studies using graph neural networks (GNNs) for image classification have focused on graphs generated from a regular grid of pixels or similar-sized superpixels. In the latter, a single target number of superpixels is defined for an entire dataset irrespective of differences across images and their intrinsic multiscale structure. On the contrary, this study investigates image classification using graphs generated from an image-specific number of multiscale superpixels. We propose WaveMesh, a new wavelet-based superpixeling algorithm, where the number and sizes of superpixels in an image are systematically computed based on its content. WaveMesh superpixel graphs are structurally different from similar-sized superpixel graphs. We use SplineCNN, a state-of-the-art network for image graph classification, to compare WaveMesh and similar-sized superpixels. Using SplineCNN, we perform extensive experiments on three benchmark datasets under three local-pooling settings: 1) no pooling, 2) GraclusPool, and 3) WavePool, a novel spatially heterogeneous pooling scheme tailored to WaveMesh superpixels. Our experiments demonstrate that SplineCNN learns from multiscale WaveMesh superpixels on-par with similar-sized superpixels. In all WaveMesh experiments, GraclusPool performs poorer than no pooling / WavePool, indicating that poor choice of pooling can result in inferior performance while learning from multiscale superpixels.
Computational model discovery with reinforcement learning
Dec 29, 2019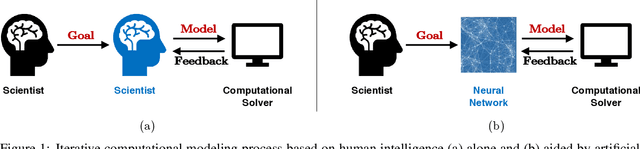
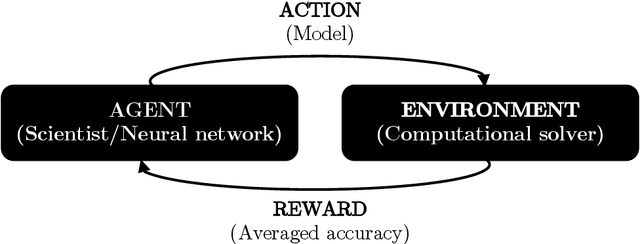
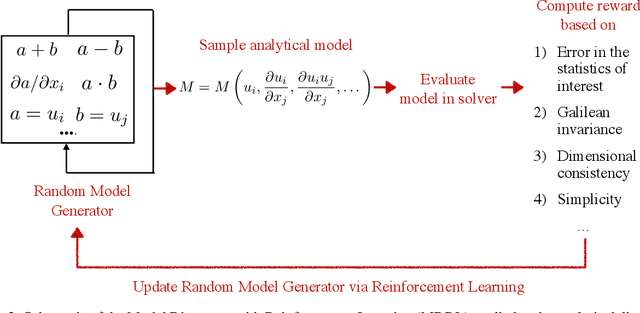

Abstract:The motivation of this study is to leverage recent breakthroughs in artificial intelligence research to unlock novel solutions to important scientific problems encountered in computational science. To address the human intelligence limitations in discovering reduced-order models, we propose to supplement human thinking with artificial intelligence. Our three-pronged strategy consists of learning (i) models expressed in analytical form, (ii) which are evaluated a posteriori, and iii) using exclusively integral quantities from the reference solution as prior knowledge. In point (i), we pursue interpretable models expressed symbolically as opposed to black-box neural networks, the latter only being used during learning to efficiently parameterize the large search space of possible models. In point (ii), learned models are dynamically evaluated a posteriori in the computational solver instead of based on a priori information from preprocessed high-fidelity data, thereby accounting for the specificity of the solver at hand such as its numerics. Finally in point (iii), the exploration of new models is solely guided by predefined integral quantities, e.g., averaged quantities of engineering interest in Reynolds-averaged or large-eddy simulations (LES). We use a coupled deep reinforcement learning framework and computational solver to concurrently achieve these objectives. The combination of reinforcement learning with objectives (i), (ii) and (iii) differentiate our work from previous modeling attempts based on machine learning. In this report, we provide a high-level description of the model discovery framework with reinforcement learning. The method is detailed for the application of discovering missing terms in differential equations. An elementary instantiation of the method is described that discovers missing terms in the Burgers' equation.
Modified U-Net with Incorporation of Object-Dependent High Level Features for Improved Liver and Liver-Tumor Segmentation in CT Images
Oct 31, 2019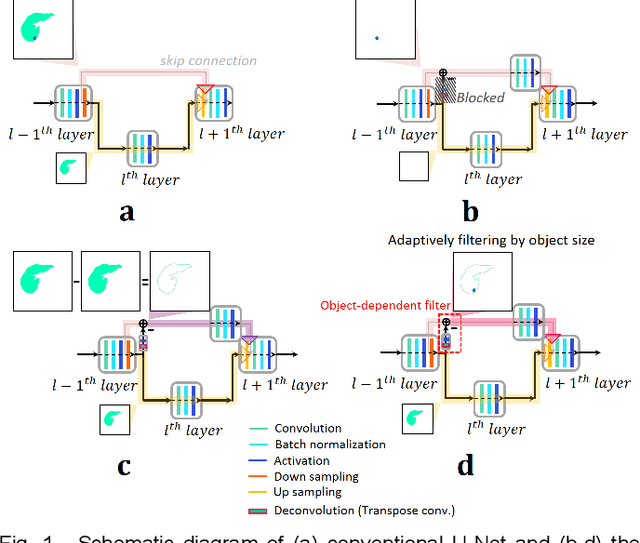
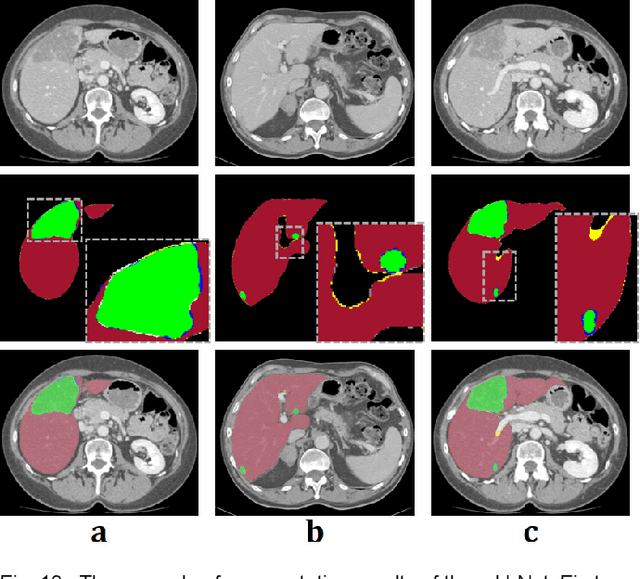
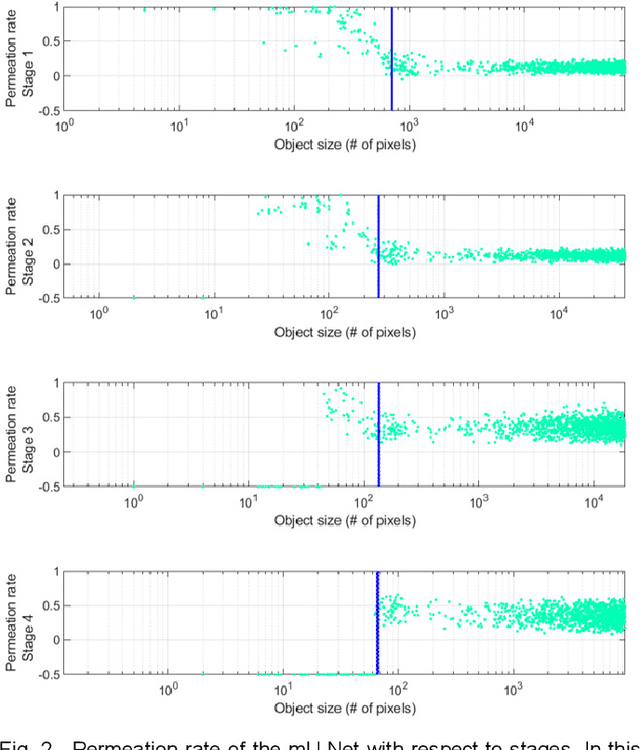

Abstract:Segmentation of livers and liver tumors is one of the most important steps in radiation therapy of hepatocellular carcinoma. The segmentation task is often done manually, making it tedious, labor intensive, and subject to intra-/inter- operator variations. While various algorithms for delineating organ-at-risks (OARs) and tumor targets have been proposed, automatic segmentation of livers and liver tumors remains intractable due to their low tissue contrast with respect to the surrounding organs and their deformable shape in CT images. The U-Net has gained increasing popularity recently for image analysis tasks and has shown promising results. Conventional U-Net architectures, however, suffer from three major drawbacks. To cope with these problems, we added a residual path with deconvolution and activation operations to the skip connection of the U-Net to avoid duplication of low resolution information of features. In the case of small object inputs, features in the skip connection are not incorporated with features in the residual path. Furthermore, the proposed architecture has additional convolution layers in the skip connection in order to extract high level global features of small object inputs as well as high level features of high resolution edge information of large object inputs. Efficacy of the modified U-Net (mU-Net) was demonstrated using the public dataset of Liver tumor segmentation (LiTS) challenge 2017. The proposed mU-Net outperformed existing state-of-art networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge