Ruoxiu Xiao
EAR: Edge-Aware Reconstruction of 3-D vertebrae structures from bi-planar X-ray images
Jul 30, 2024Abstract:X-ray images ease the diagnosis and treatment process due to their rapid imaging speed and high resolution. However, due to the projection process of X-ray imaging, much spatial information has been lost. To accurately provide efficient spinal morphological and structural information, reconstructing the 3-D structures of the spine from the 2-D X-ray images is essential. It is challenging for current reconstruction methods to preserve the edge information and local shapes of the asymmetrical vertebrae structures. In this study, we propose a new Edge-Aware Reconstruction network (EAR) to focus on the performance improvement of the edge information and vertebrae shapes. In our network, by using the auto-encoder architecture as the backbone, the edge attention module and frequency enhancement module are proposed to strengthen the perception of the edge reconstruction. Meanwhile, we also combine four loss terms, including reconstruction loss, edge loss, frequency loss and projection loss. The proposed method is evaluated using three publicly accessible datasets and compared with four state-of-the-art models. The proposed method is superior to other methods and achieves 25.32%, 15.32%, 86.44%, 80.13%, 23.7612 and 0.3014 with regard to MSE, MAE, Dice, SSIM, PSNR and frequency distance. Due to the end-to-end and accurate reconstruction process, EAR can provide sufficient 3-D spatial information and precise preoperative surgical planning guidance.
Multi-view X-ray Image Synthesis with Multiple Domain Disentanglement from CT Scans
Apr 18, 2024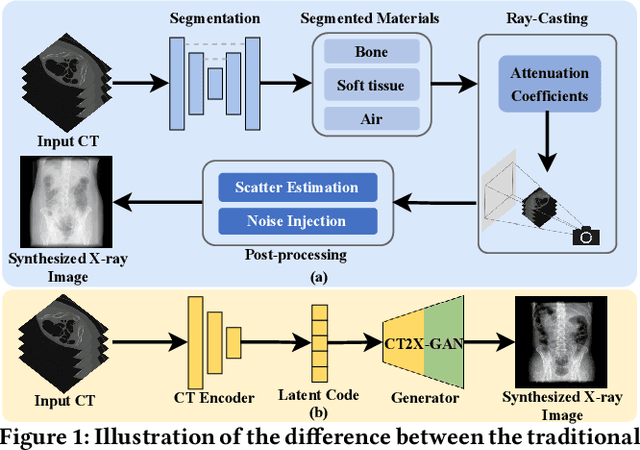

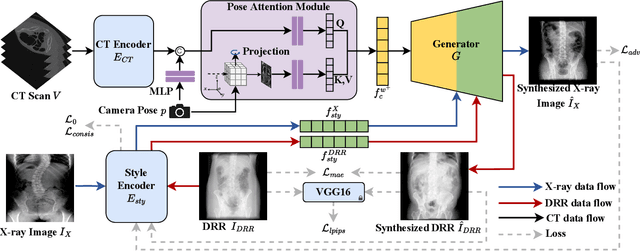
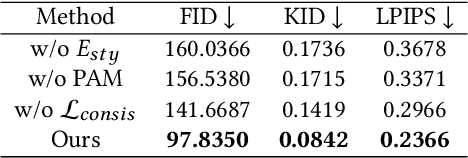
Abstract:X-ray images play a vital role in the intraoperative processes due to their high resolution and fast imaging speed and greatly promote the subsequent segmentation, registration and reconstruction. However, over-dosed X-rays superimpose potential risks to human health to some extent. Data-driven algorithms from volume scans to X-ray images are restricted by the scarcity of paired X-ray and volume data. Existing methods are mainly realized by modelling the whole X-ray imaging procedure. In this study, we propose a learning-based approach termed CT2X-GAN to synthesize the X-ray images in an end-to-end manner using the content and style disentanglement from three different image domains. Our method decouples the anatomical structure information from CT scans and style information from unpaired real X-ray images/ digital reconstructed radiography (DRR) images via a series of decoupling encoders. Additionally, we introduce a novel consistency regularization term to improve the stylistic resemblance between synthesized X-ray images and real X-ray images. Meanwhile, we also impose a supervised process by computing the similarity of computed real DRR and synthesized DRR images. We further develop a pose attention module to fully strengthen the comprehensive information in the decoupled content code from CT scans, facilitating high-quality multi-view image synthesis in the lower 2D space. Extensive experiments were conducted on the publicly available CTSpine1K dataset and achieved 97.8350, 0.0842 and 3.0938 in terms of FID, KID and defined user-scored X-ray similarity, respectively. In comparison with 3D-aware methods ($\pi$-GAN, EG3D), CT2X-GAN is superior in improving the synthesis quality and realistic to the real X-ray images.
Degradation-invariant Enhancement of Fundus Images via Pyramid Constraint Network
Oct 18, 2022Abstract:As an economical and efficient fundus imaging modality, retinal fundus images have been widely adopted in clinical fundus examination. Unfortunately, fundus images often suffer from quality degradation caused by imaging interferences, leading to misdiagnosis. Despite impressive enhancement performances that state-of-the-art methods have achieved, challenges remain in clinical scenarios. For boosting the clinical deployment of fundus image enhancement, this paper proposes the pyramid constraint to develop a degradation-invariant enhancement network (PCE-Net), which mitigates the demand for clinical data and stably enhances unknown data. Firstly, high-quality images are randomly degraded to form sequences of low-quality ones sharing the same content (SeqLCs). Then individual low-quality images are decomposed to Laplacian pyramid features (LPF) as the multi-level input for the enhancement. Subsequently, a feature pyramid constraint (FPC) for the sequence is introduced to enforce the PCE-Net to learn a degradation-invariant model. Extensive experiments have been conducted under the evaluation metrics of enhancement and segmentation. The effectiveness of the PCE-Net was demonstrated in comparison with state-of-the-art methods and the ablation study. The source code of this study is publicly available at https://github.com/HeverLaw/PCENet-Image-Enhancement.
Machine Learning Techniques for Biomedical Image Segmentation: An Overview of Technical Aspects and Introduction to State-of-Art Applications
Nov 06, 2019Abstract:In recent years, significant progress has been made in developing more accurate and efficient machine learning algorithms for segmentation of medical and natural images. In this review article, we highlight the imperative role of machine learning algorithms in enabling efficient and accurate segmentation in the field of medical imaging. We specifically focus on several key studies pertaining to the application of machine learning methods to biomedical image segmentation. We review classical machine learning algorithms such as Markov random fields, k-means clustering, random forest, etc. Although such classical learning models are often less accurate compared to the deep learning techniques, they are often more sample efficient and have a less complex structure. We also review different deep learning architectures, such as the artificial neural networks (ANNs), the convolutional neural networks (CNNs), and the recurrent neural networks (RNNs), and present the segmentation results attained by those learning models that were published in the past three years. We highlight the successes and limitations of each machine learning paradigm. In addition, we discuss several challenges related to the training of different machine learning models, and we present some heuristics to address those challenges.
Modified U-Net with Incorporation of Object-Dependent High Level Features for Improved Liver and Liver-Tumor Segmentation in CT Images
Oct 31, 2019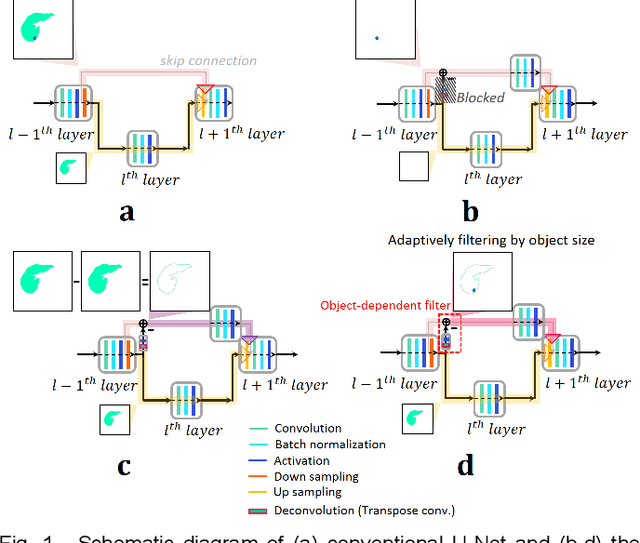
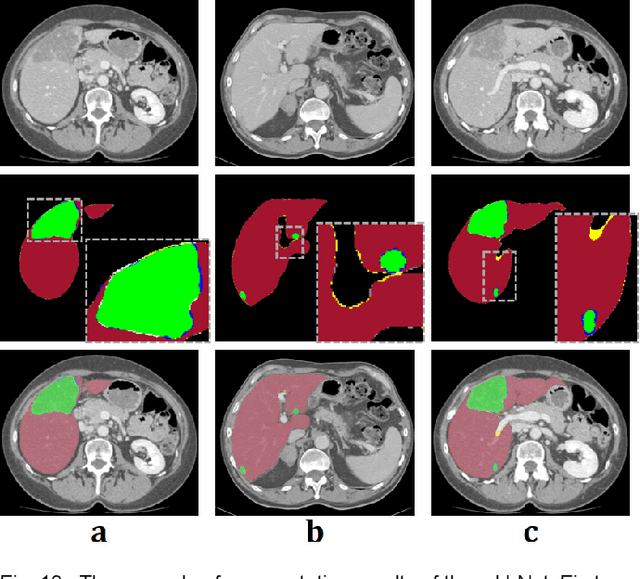
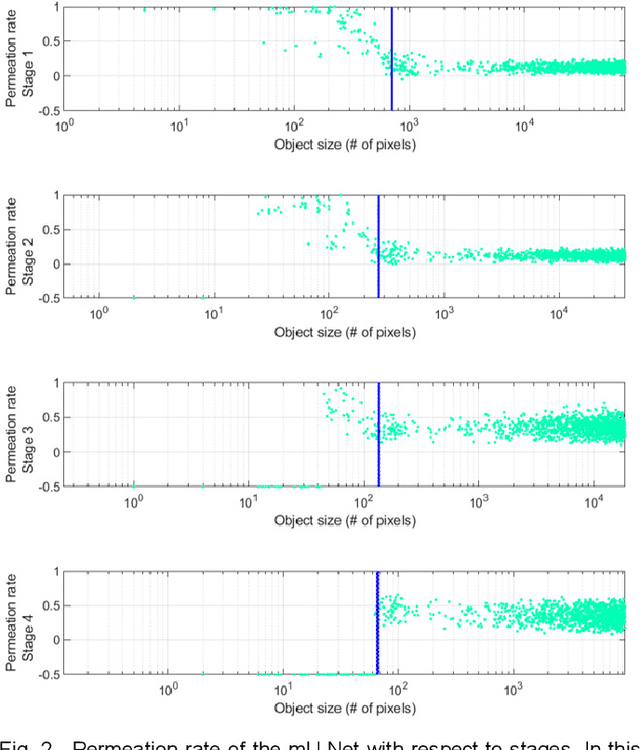

Abstract:Segmentation of livers and liver tumors is one of the most important steps in radiation therapy of hepatocellular carcinoma. The segmentation task is often done manually, making it tedious, labor intensive, and subject to intra-/inter- operator variations. While various algorithms for delineating organ-at-risks (OARs) and tumor targets have been proposed, automatic segmentation of livers and liver tumors remains intractable due to their low tissue contrast with respect to the surrounding organs and their deformable shape in CT images. The U-Net has gained increasing popularity recently for image analysis tasks and has shown promising results. Conventional U-Net architectures, however, suffer from three major drawbacks. To cope with these problems, we added a residual path with deconvolution and activation operations to the skip connection of the U-Net to avoid duplication of low resolution information of features. In the case of small object inputs, features in the skip connection are not incorporated with features in the residual path. Furthermore, the proposed architecture has additional convolution layers in the skip connection in order to extract high level global features of small object inputs as well as high level features of high resolution edge information of large object inputs. Efficacy of the modified U-Net (mU-Net) was demonstrated using the public dataset of Liver tumor segmentation (LiTS) challenge 2017. The proposed mU-Net outperformed existing state-of-art networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge