Hyunseok Seo
Spectrum Translation for Refinement of Image Generation (STIG) Based on Contrastive Learning and Spectral Filter Profile
Mar 08, 2024



Abstract:Currently, image generation and synthesis have remarkably progressed with generative models. Despite photo-realistic results, intrinsic discrepancies are still observed in the frequency domain. The spectral discrepancy appeared not only in generative adversarial networks but in diffusion models. In this study, we propose a framework to effectively mitigate the disparity in frequency domain of the generated images to improve generative performance of both GAN and diffusion models. This is realized by spectrum translation for the refinement of image generation (STIG) based on contrastive learning. We adopt theoretical logic of frequency components in various generative networks. The key idea, here, is to refine the spectrum of the generated image via the concept of image-to-image translation and contrastive learning in terms of digital signal processing. We evaluate our framework across eight fake image datasets and various cutting-edge models to demonstrate the effectiveness of STIG. Our framework outperforms other cutting-edges showing significant decreases in FID and log frequency distance of spectrum. We further emphasize that STIG improves image quality by decreasing the spectral anomaly. Additionally, validation results present that the frequency-based deepfake detector confuses more in the case where fake spectrums are manipulated by STIG.
Attention Guided CAM: Visual Explanations of Vision Transformer Guided by Self-Attention
Feb 07, 2024



Abstract:Vision Transformer(ViT) is one of the most widely used models in the computer vision field with its great performance on various tasks. In order to fully utilize the ViT-based architecture in various applications, proper visualization methods with a decent localization performance are necessary, but these methods employed in CNN-based models are still not available in ViT due to its unique structure. In this work, we propose an attention-guided visualization method applied to ViT that provides a high-level semantic explanation for its decision. Our method selectively aggregates the gradients directly propagated from the classification output to each self-attention, collecting the contribution of image features extracted from each location of the input image. These gradients are additionally guided by the normalized self-attention scores, which are the pairwise patch correlation scores. They are used to supplement the gradients on the patch-level context information efficiently detected by the self-attention mechanism. This approach of our method provides elaborate high-level semantic explanations with great localization performance only with the class labels. As a result, our method outperforms the previous leading explainability methods of ViT in the weakly-supervised localization task and presents great capability in capturing the full instances of the target class object. Meanwhile, our method provides a visualization that faithfully explains the model, which is demonstrated in the perturbation comparison test.
Atlas Based Segmentations via Semi-Supervised Diffeomorphic Registrations
Nov 23, 2019



Abstract:Purpose: Segmentation of organs-at-risk (OARs) is a bottleneck in current radiation oncology pipelines and is often time consuming and labor intensive. In this paper, we propose an atlas-based semi-supervised registration algorithm to generate accurate segmentations of OARs for which there are ground truth contours and rough segmentations of all other OARs in the atlas. To the best of our knowledge, this is the first study to use learning-based registration methods for the segmentation of head and neck patients and demonstrate its utility in clinical applications. Methods: Our algorithm cascades rigid and deformable deformation blocks, and takes on an atlas image (M), set of atlas-space segmentations (S_A), and a patient image (F) as inputs, while outputting patient-space segmentations of all OARs defined on the atlas. We train our model on 475 CT images taken from public archives and Stanford RadOnc Clinic (SROC), validate on 5 CT images from SROC, and test our model on 20 CT images from SROC. Results: Our method outperforms current state of the art learning-based registration algorithms and achieves an overall dice score of 0.789 on our test set. Moreover, our method yields a performance comparable to manual segmentation and supervised segmentation, while solving a much more complex registration problem. Whereas supervised segmentation methods only automate the segmentation process for a select few number of OARs, we demonstrate that our methods can achieve similar performance for OARs of interest, while also providing segmentations for every other OAR on the provided atlas. Conclusions: Our proposed algorithm has significant clinical applications and could help reduce the bottleneck for segmentation of head and neck OARs. Further, our results demonstrate that semi-supervised diffeomorphic registration can be accurately applied to both registration and segmentation problems.
Machine Learning Techniques for Biomedical Image Segmentation: An Overview of Technical Aspects and Introduction to State-of-Art Applications
Nov 06, 2019Abstract:In recent years, significant progress has been made in developing more accurate and efficient machine learning algorithms for segmentation of medical and natural images. In this review article, we highlight the imperative role of machine learning algorithms in enabling efficient and accurate segmentation in the field of medical imaging. We specifically focus on several key studies pertaining to the application of machine learning methods to biomedical image segmentation. We review classical machine learning algorithms such as Markov random fields, k-means clustering, random forest, etc. Although such classical learning models are often less accurate compared to the deep learning techniques, they are often more sample efficient and have a less complex structure. We also review different deep learning architectures, such as the artificial neural networks (ANNs), the convolutional neural networks (CNNs), and the recurrent neural networks (RNNs), and present the segmentation results attained by those learning models that were published in the past three years. We highlight the successes and limitations of each machine learning paradigm. In addition, we discuss several challenges related to the training of different machine learning models, and we present some heuristics to address those challenges.
Modified U-Net with Incorporation of Object-Dependent High Level Features for Improved Liver and Liver-Tumor Segmentation in CT Images
Oct 31, 2019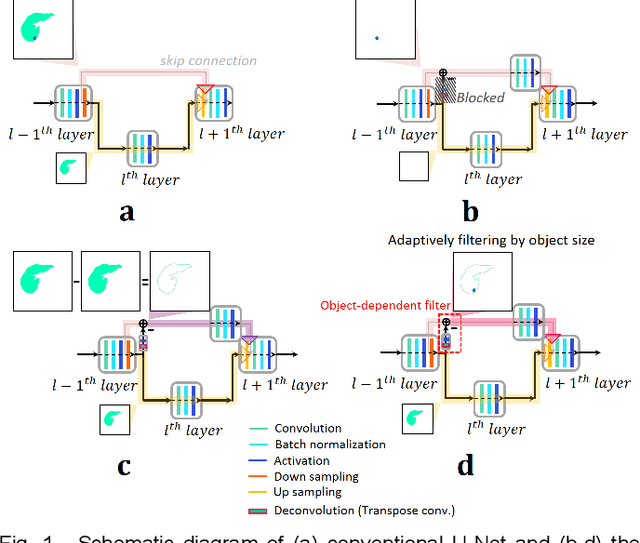
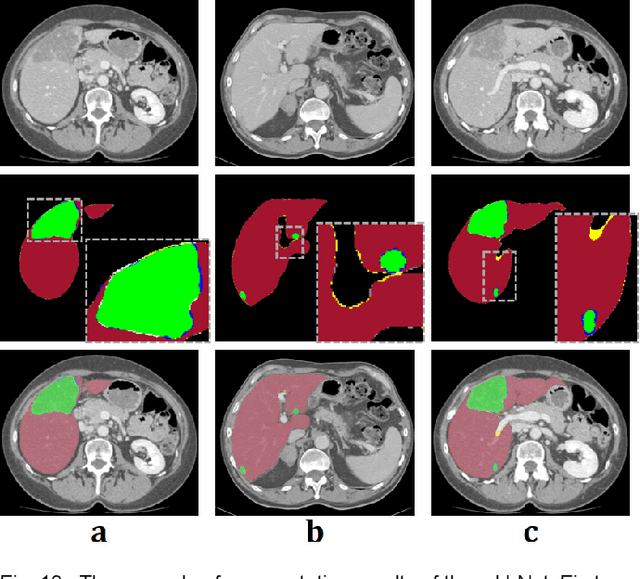
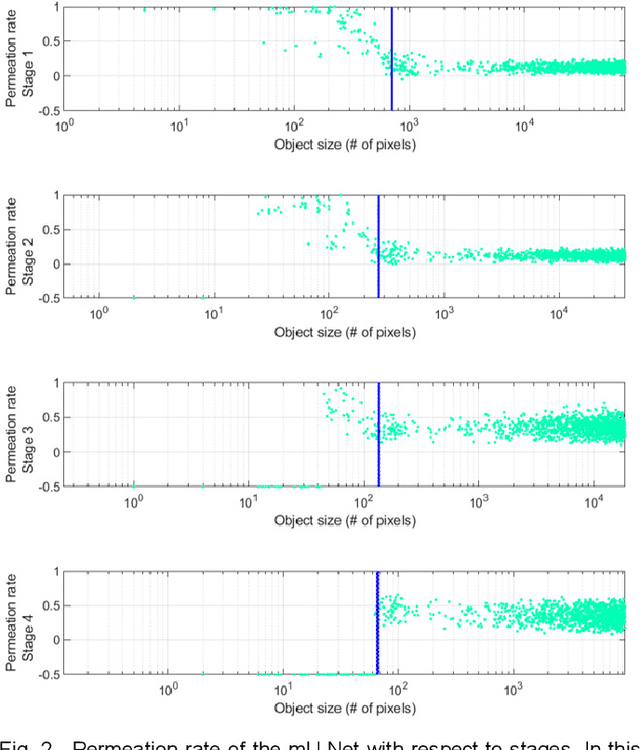

Abstract:Segmentation of livers and liver tumors is one of the most important steps in radiation therapy of hepatocellular carcinoma. The segmentation task is often done manually, making it tedious, labor intensive, and subject to intra-/inter- operator variations. While various algorithms for delineating organ-at-risks (OARs) and tumor targets have been proposed, automatic segmentation of livers and liver tumors remains intractable due to their low tissue contrast with respect to the surrounding organs and their deformable shape in CT images. The U-Net has gained increasing popularity recently for image analysis tasks and has shown promising results. Conventional U-Net architectures, however, suffer from three major drawbacks. To cope with these problems, we added a residual path with deconvolution and activation operations to the skip connection of the U-Net to avoid duplication of low resolution information of features. In the case of small object inputs, features in the skip connection are not incorporated with features in the residual path. Furthermore, the proposed architecture has additional convolution layers in the skip connection in order to extract high level global features of small object inputs as well as high level features of high resolution edge information of large object inputs. Efficacy of the modified U-Net (mU-Net) was demonstrated using the public dataset of Liver tumor segmentation (LiTS) challenge 2017. The proposed mU-Net outperformed existing state-of-art networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge