Varun Vasudevan
To See or To Read: User Behavior Reasoning in Multimodal LLMs
Nov 05, 2025Abstract:Multimodal Large Language Models (MLLMs) are reshaping how modern agentic systems reason over sequential user-behavior data. However, whether textual or image representations of user behavior data are more effective for maximizing MLLM performance remains underexplored. We present \texttt{BehaviorLens}, a systematic benchmarking framework for assessing modality trade-offs in user-behavior reasoning across six MLLMs by representing transaction data as (1) a text paragraph, (2) a scatter plot, and (3) a flowchart. Using a real-world purchase-sequence dataset, we find that when data is represented as images, MLLMs next-purchase prediction accuracy is improved by 87.5% compared with an equivalent textual representation without any additional computational cost.
LLM-driven Constrained Copy Generation through Iterative Refinement
Apr 14, 2025Abstract:Crafting a marketing message (copy), or copywriting is a challenging generation task, as the copy must adhere to various constraints. Copy creation is inherently iterative for humans, starting with an initial draft followed by successive refinements. However, manual copy creation is time-consuming and expensive, resulting in only a few copies for each use case. This limitation restricts our ability to personalize content to customers. Contrary to the manual approach, LLMs can generate copies quickly, but the generated content does not consistently meet all the constraints on the first attempt (similar to humans). While recent studies have shown promise in improving constrained generation through iterative refinement, they have primarily addressed tasks with only a few simple constraints. Consequently, the effectiveness of iterative refinement for tasks such as copy generation, which involves many intricate constraints, remains unclear. To address this gap, we propose an LLM-based end-to-end framework for scalable copy generation using iterative refinement. To the best of our knowledge, this is the first study to address multiple challenging constraints simultaneously in copy generation. Examples of these constraints include length, topics, keywords, preferred lexical ordering, and tone of voice. We demonstrate the performance of our framework by creating copies for e-commerce banners for three different use cases of varying complexity. Our results show that iterative refinement increases the copy success rate by $16.25-35.91$% across use cases. Furthermore, the copies generated using our approach outperformed manually created content in multiple pilot studies using a multi-armed bandit framework. The winning copy improved the click-through rate by $38.5-45.21$%.
Learning Treatment Plan Representations for Content Based Image Retrieval
Jun 06, 2022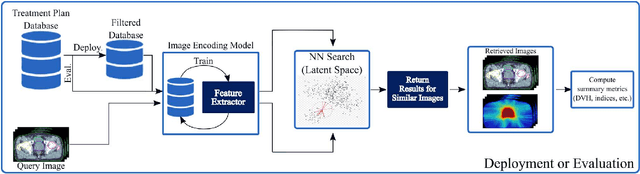
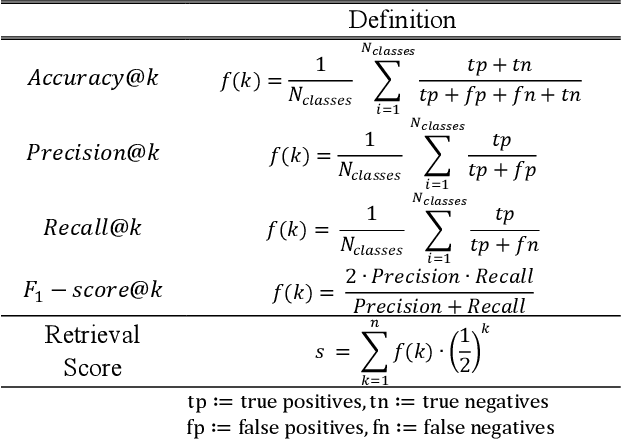
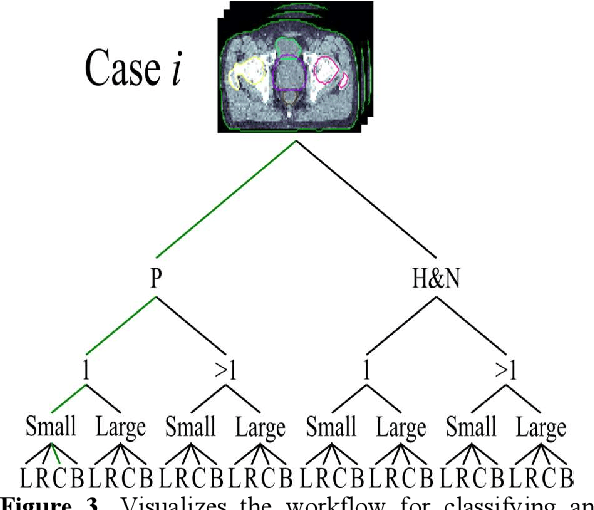
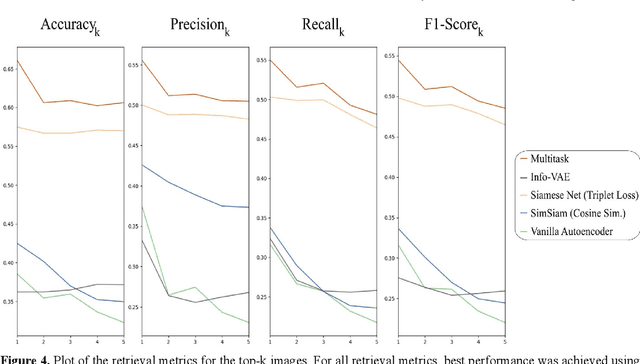
Abstract:Objective: Knowledge based planning (KBP) typically involves training an end-to-end deep learning model to predict dose distributions. However, training end-to-end KBP methods may be associated with practical limitations due to the limited size of medical datasets that are often used. To address these limitations, we propose a content based image retrieval (CBIR) method for retrieving dose distributions of previously planned patients based on anatomical similarity. Approach: Our proposed CBIR method trains a representation model that produces latent space embeddings of a patient's anatomical information. The latent space embeddings of new patients are then compared against those of previous patients in a database for image retrieval of dose distributions. Summary metrics (e.g. dose-volume histogram, conformity index, homogeneity index, etc.) are computed and can then be utilized in subsequent automated planning. All source code for this project is available on github. Main Results: The retrieval performance of various CBIR methods is evaluated on a dataset consisting of both publicly available plans and clinical plans from our institution. This study compares various encoding methods, ranging from simple autoencoders to more recent Siamese networks like SimSiam, and the best performance was observed for the multitask Siamese network. Significance: Applying CBIR to inform subsequent treatment planning potentially addresses many limitations associated with end-to-end KBP. Our current results demonstrate that excellent image retrieval performance can be obtained through slight changes to previously developed Siamese networks. We hope to integrate CBIR into automated planning workflow in future works, potentially through methods like the MetaPlanner framework.
Image Classification using Graph Neural Network and Multiscale Wavelet Superpixels
Jan 29, 2022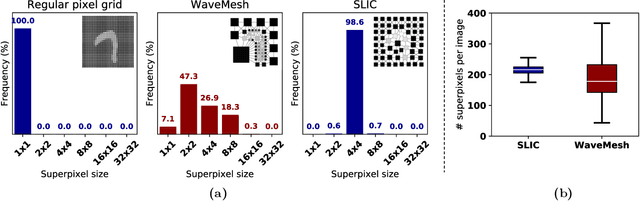
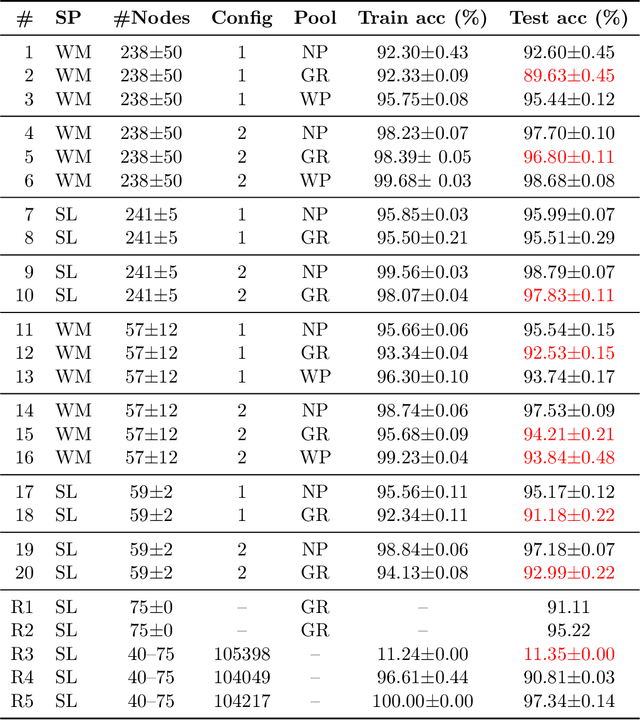
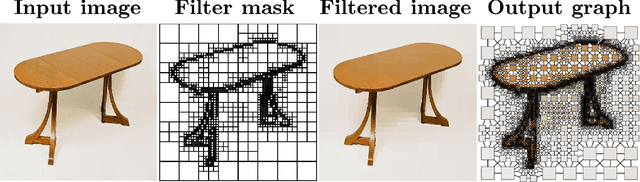
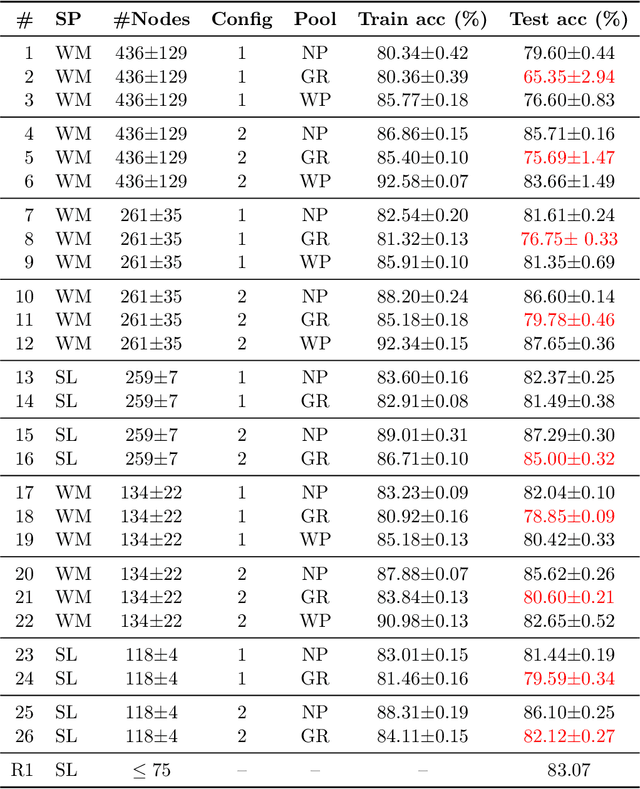
Abstract:Prior studies using graph neural networks (GNNs) for image classification have focused on graphs generated from a regular grid of pixels or similar-sized superpixels. In the latter, a single target number of superpixels is defined for an entire dataset irrespective of differences across images and their intrinsic multiscale structure. On the contrary, this study investigates image classification using graphs generated from an image-specific number of multiscale superpixels. We propose WaveMesh, a new wavelet-based superpixeling algorithm, where the number and sizes of superpixels in an image are systematically computed based on its content. WaveMesh superpixel graphs are structurally different from similar-sized superpixel graphs. We use SplineCNN, a state-of-the-art network for image graph classification, to compare WaveMesh and similar-sized superpixels. Using SplineCNN, we perform extensive experiments on three benchmark datasets under three local-pooling settings: 1) no pooling, 2) GraclusPool, and 3) WavePool, a novel spatially heterogeneous pooling scheme tailored to WaveMesh superpixels. Our experiments demonstrate that SplineCNN learns from multiscale WaveMesh superpixels on-par with similar-sized superpixels. In all WaveMesh experiments, GraclusPool performs poorer than no pooling / WavePool, indicating that poor choice of pooling can result in inferior performance while learning from multiscale superpixels.
Machine Learning Techniques for Biomedical Image Segmentation: An Overview of Technical Aspects and Introduction to State-of-Art Applications
Nov 06, 2019Abstract:In recent years, significant progress has been made in developing more accurate and efficient machine learning algorithms for segmentation of medical and natural images. In this review article, we highlight the imperative role of machine learning algorithms in enabling efficient and accurate segmentation in the field of medical imaging. We specifically focus on several key studies pertaining to the application of machine learning methods to biomedical image segmentation. We review classical machine learning algorithms such as Markov random fields, k-means clustering, random forest, etc. Although such classical learning models are often less accurate compared to the deep learning techniques, they are often more sample efficient and have a less complex structure. We also review different deep learning architectures, such as the artificial neural networks (ANNs), the convolutional neural networks (CNNs), and the recurrent neural networks (RNNs), and present the segmentation results attained by those learning models that were published in the past three years. We highlight the successes and limitations of each machine learning paradigm. In addition, we discuss several challenges related to the training of different machine learning models, and we present some heuristics to address those challenges.
On Sample Complexity of Projection-Free Primal-Dual Methods for Learning Mixture Policies in Markov Decision Processes
Mar 20, 2019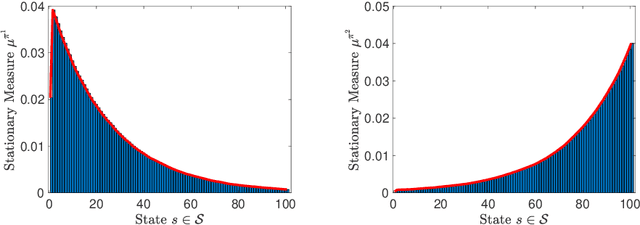
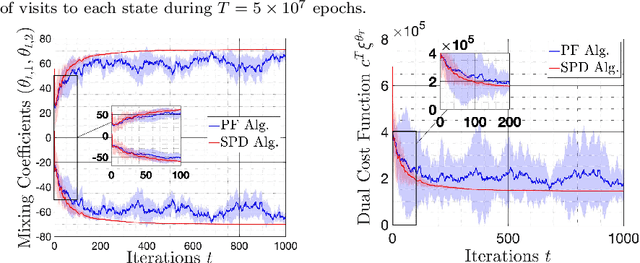
Abstract:We study the problem of learning policy of an infinite-horizon, discounted cost, Markov decision process (MDP) with a large number of states. We compute the actions of a policy that is nearly as good as a policy chosen by a suitable oracle from a given mixture policy class characterized by the convex hull of a set of known base policies. To learn the coefficients of the mixture model, we recast the problem as an approximate linear programming (ALP) formulation for MDPs, where the feature vectors correspond to the occupation measures of the base policies defined on the state-action space. We then propose a projection-free stochastic primal-dual method with the Bregman divergence to solve the characterized ALP. Furthermore, we analyze the probably approximately correct (PAC) sample complexity of the proposed stochastic algorithm, namely the number of queries required to achieve near optimal objective value. We also propose a modification of our proposed algorithm with the polytope constraint sampling for the smoothed ALP, where the restriction to lower bounding approximations are relaxed. In addition, we apply the proposed algorithms to a queuing problem, and compare their performance with a penalty function algorithm. The numerical results illustrates that the primal-dual achieves better efficiency and low variance across different trials compared to the penalty function method.
Multiple Kernel Learning from $U$-Statistics of Empirical Measures in the Feature Space
Feb 27, 2019


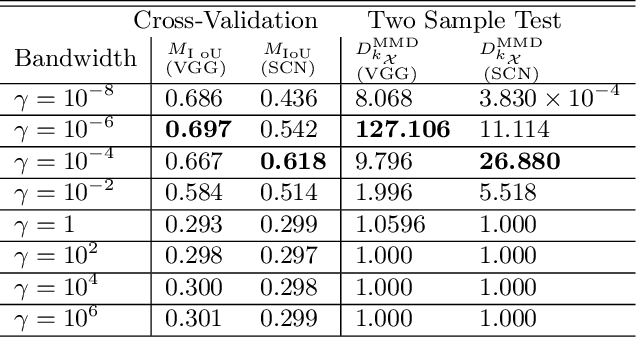
Abstract:We propose a novel data-driven method to learn multiple kernels in kernel methods of statistical machine learning from training samples. The proposed kernel learning algorithm is based on a $U$-statistics of the empirical marginal distributions of features in the feature space given their class labels. We prove the consistency of the $U$-statistic estimate using the empirical distributions for kernel learning. In particular, we show that the empirical estimate of $U$-statistic converges to its population value with respect to all admissible distributions as the number of the training samples increase. We also prove the sample optimality of the estimate by establishing a minimax lower bound via Fano's method. In addition, we establish the generalization bounds of the proposed kernel learning approach by computing novel upper bounds on the Rademacher and Gaussian complexities using the concentration of measures for the quadratic matrix forms.We apply the proposed kernel learning approach to classification of the real-world data-sets using the kernel SVM and compare the results with $5$-fold cross-validation for the kernel model selection problem. We also apply the proposed kernel learning approach to devise novel architectures for the semantic segmentation of biomedical images. The proposed segmentation networks are suited for training on small data-sets and employ new mechanisms to generate representations from input images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge