Hongyi Ren
Metal Artifact Reduction in 2D CT Images with Self-supervised Cross-domain Learning
Sep 28, 2021Abstract:The presence of metallic implants often introduces severe metal artifacts in the X-ray CT images, which could adversely influence clinical diagnosis or dose calculation in radiation therapy. In this work, we present a novel deep-learning-based approach for metal artifact reduction (MAR). In order to alleviate the need for anatomically identical CT image pairs (i.e., metal artifact-corrupted CT image and metal artifact-free CT image) for network learning, we propose a self-supervised cross-domain learning framework. Specifically, we train a neural network to restore the metal trace region values in the given metal-free sinogram, where the metal trace is identified by the forward projection of metal masks. We then design a novel FBP reconstruction loss to encourage the network to generate more perfect completion results and a residual-learning-based image refinement module to reduce the secondary artifacts in the reconstructed CT images. To preserve the fine structure details and fidelity of the final MAR image, instead of directly adopting CNN-refined images as output, we incorporate the metal trace replacement into our framework and replace the metal-affected projections of the original sinogram with the prior sinogram generated by the forward projection of the CNN output. We then use the filtered backward projection (FBP) algorithms for final MAR image reconstruction. We conduct an extensive evaluation on simulated and real artifact data to show the effectiveness of our design. Our method produces superior MAR results and outperforms other compelling methods. We also demonstrate the potential of our framework for other organ sites.
Machine Learning Techniques for Biomedical Image Segmentation: An Overview of Technical Aspects and Introduction to State-of-Art Applications
Nov 06, 2019Abstract:In recent years, significant progress has been made in developing more accurate and efficient machine learning algorithms for segmentation of medical and natural images. In this review article, we highlight the imperative role of machine learning algorithms in enabling efficient and accurate segmentation in the field of medical imaging. We specifically focus on several key studies pertaining to the application of machine learning methods to biomedical image segmentation. We review classical machine learning algorithms such as Markov random fields, k-means clustering, random forest, etc. Although such classical learning models are often less accurate compared to the deep learning techniques, they are often more sample efficient and have a less complex structure. We also review different deep learning architectures, such as the artificial neural networks (ANNs), the convolutional neural networks (CNNs), and the recurrent neural networks (RNNs), and present the segmentation results attained by those learning models that were published in the past three years. We highlight the successes and limitations of each machine learning paradigm. In addition, we discuss several challenges related to the training of different machine learning models, and we present some heuristics to address those challenges.
On Sample Complexity of Projection-Free Primal-Dual Methods for Learning Mixture Policies in Markov Decision Processes
Mar 20, 2019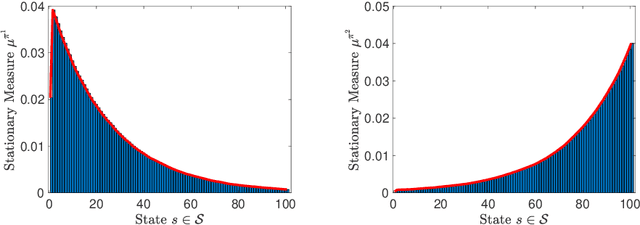
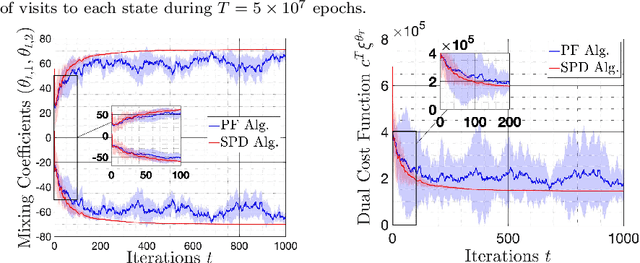
Abstract:We study the problem of learning policy of an infinite-horizon, discounted cost, Markov decision process (MDP) with a large number of states. We compute the actions of a policy that is nearly as good as a policy chosen by a suitable oracle from a given mixture policy class characterized by the convex hull of a set of known base policies. To learn the coefficients of the mixture model, we recast the problem as an approximate linear programming (ALP) formulation for MDPs, where the feature vectors correspond to the occupation measures of the base policies defined on the state-action space. We then propose a projection-free stochastic primal-dual method with the Bregman divergence to solve the characterized ALP. Furthermore, we analyze the probably approximately correct (PAC) sample complexity of the proposed stochastic algorithm, namely the number of queries required to achieve near optimal objective value. We also propose a modification of our proposed algorithm with the polytope constraint sampling for the smoothed ALP, where the restriction to lower bounding approximations are relaxed. In addition, we apply the proposed algorithms to a queuing problem, and compare their performance with a penalty function algorithm. The numerical results illustrates that the primal-dual achieves better efficiency and low variance across different trials compared to the penalty function method.
Multiple Kernel Learning from $U$-Statistics of Empirical Measures in the Feature Space
Feb 27, 2019


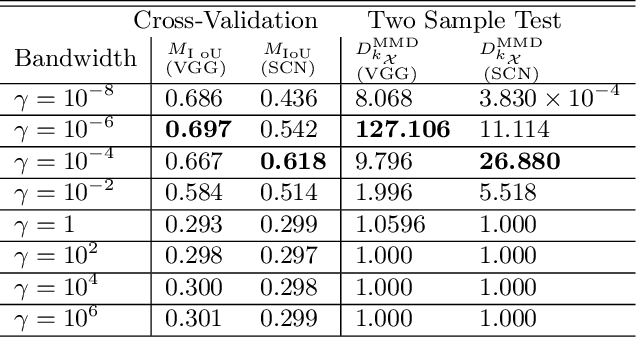
Abstract:We propose a novel data-driven method to learn multiple kernels in kernel methods of statistical machine learning from training samples. The proposed kernel learning algorithm is based on a $U$-statistics of the empirical marginal distributions of features in the feature space given their class labels. We prove the consistency of the $U$-statistic estimate using the empirical distributions for kernel learning. In particular, we show that the empirical estimate of $U$-statistic converges to its population value with respect to all admissible distributions as the number of the training samples increase. We also prove the sample optimality of the estimate by establishing a minimax lower bound via Fano's method. In addition, we establish the generalization bounds of the proposed kernel learning approach by computing novel upper bounds on the Rademacher and Gaussian complexities using the concentration of measures for the quadratic matrix forms.We apply the proposed kernel learning approach to classification of the real-world data-sets using the kernel SVM and compare the results with $5$-fold cross-validation for the kernel model selection problem. We also apply the proposed kernel learning approach to devise novel architectures for the semantic segmentation of biomedical images. The proposed segmentation networks are suited for training on small data-sets and employ new mechanisms to generate representations from input images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge