Mengyu Jia
Quasi-multimodal-based pathophysiological feature learning for retinal disease diagnosis
Feb 03, 2026Abstract:Retinal diseases spanning a broad spectrum can be effectively identified and diagnosed using complementary signals from multimodal data. However, multimodal diagnosis in ophthalmic practice is typically challenged in terms of data heterogeneity, potential invasiveness, registration complexity, and so on. As such, a unified framework that integrates multimodal data synthesis and fusion is proposed for retinal disease classification and grading. Specifically, the synthesized multimodal data incorporates fundus fluorescein angiography (FFA), multispectral imaging (MSI), and saliency maps that emphasize latent lesions as well as optic disc/cup regions. Parallel models are independently trained to learn modality-specific representations that capture cross-pathophysiological signatures. These features are then adaptively calibrated within and across modalities to perform information pruning and flexible integration according to downstream tasks. The proposed learning system is thoroughly interpreted through visualizations in both image and feature spaces. Extensive experiments on two public datasets demonstrated the superiority of our approach over state-of-the-art ones in the tasks of multi-label classification (F1-score: 0.683, AUC: 0.953) and diabetic retinopathy grading (Accuracy:0.842, Kappa: 0.861). This work not only enhances the accuracy and efficiency of retinal disease screening but also offers a scalable framework for data augmentation across various medical imaging modalities.
Representation Learning on Heterostructures via Heterogeneous Anonymous Walks
Jan 18, 2022
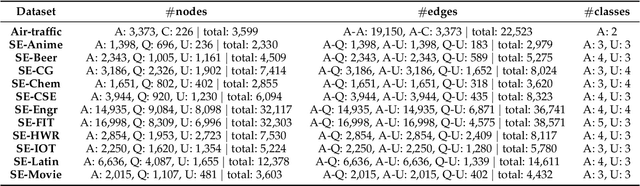
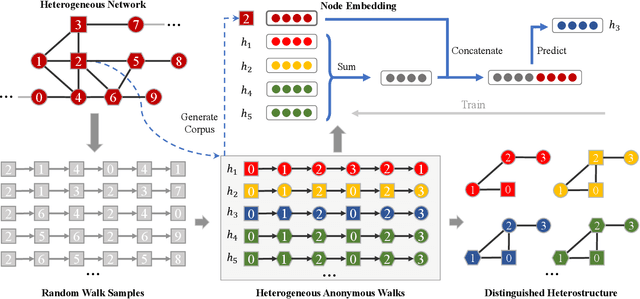
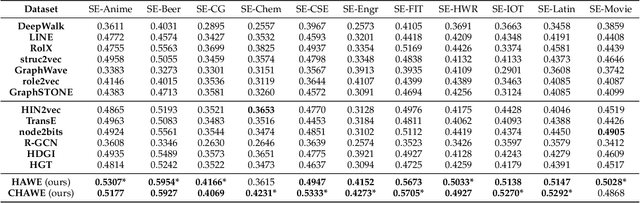
Abstract:Capturing structural similarity has been a hot topic in the field of network embedding recently due to its great help in understanding the node functions and behaviors. However, existing works have paid very much attention to learning structures on homogeneous networks while the related study on heterogeneous networks is still a void. In this paper, we try to take the first step for representation learning on heterostructures, which is very challenging due to their highly diverse combinations of node types and underlying structures. To effectively distinguish diverse heterostructures, we firstly propose a theoretically guaranteed technique called heterogeneous anonymous walk (HAW) and its variant coarse HAW (CHAW). Then, we devise the heterogeneous anonymous walk embedding (HAWE) and its variant coarse HAWE in a data-driven manner to circumvent using an extremely large number of possible walks and train embeddings by predicting occurring walks in the neighborhood of each node. Finally, we design and apply extensive and illustrative experiments on synthetic and real-world networks to build a benchmark on heterostructure learning and evaluate the effectiveness of our methods. The results demonstrate our methods achieve outstanding performance compared with both homogeneous and heterogeneous classic methods, and can be applied on large-scale networks.
Self-supervised Feature Learning via Exploiting Multi-modal Data for Retinal Disease Diagnosis
Jul 21, 2020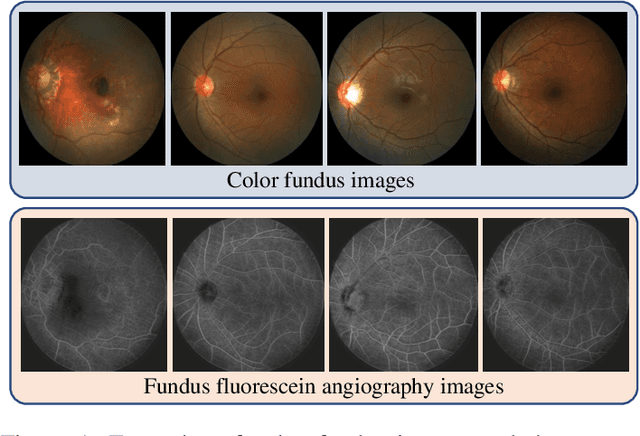
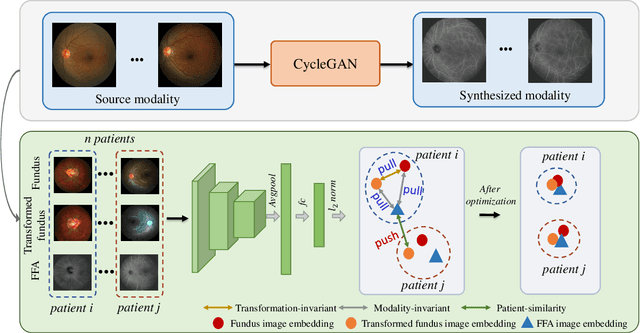
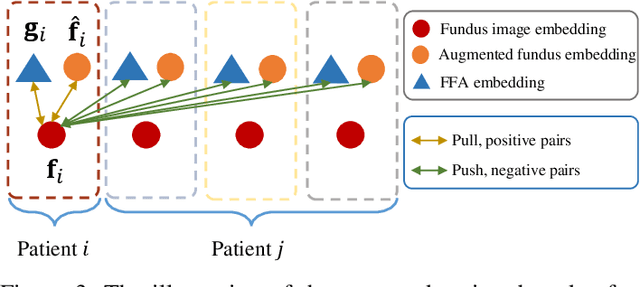
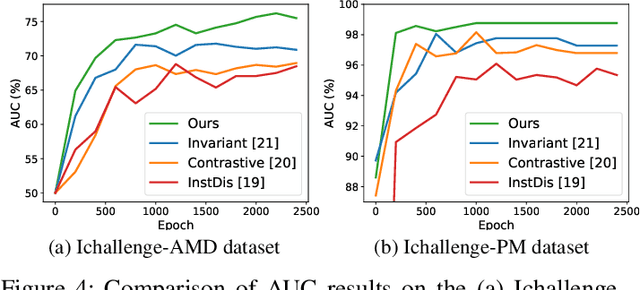
Abstract:The automatic diagnosis of various retinal diseases from fundus images is important to support clinical decision-making. However, developing such automatic solutions is challenging due to the requirement of a large amount of human-annotated data. Recently, unsupervised/self-supervised feature learning techniques receive a lot of attention, as they do not need massive annotations. Most of the current self-supervised methods are analyzed with single imaging modality and there is no method currently utilize multi-modal images for better results. Considering that the diagnostics of various vitreoretinal diseases can greatly benefit from another imaging modality, e.g., FFA, this paper presents a novel self-supervised feature learning method by effectively exploiting multi-modal data for retinal disease diagnosis. To achieve this, we first synthesize the corresponding FFA modality and then formulate a patient feature-based softmax embedding objective. Our objective learns both modality-invariant features and patient-similarity features. Through this mechanism, the neural network captures the semantically shared information across different modalities and the apparent visual similarity between patients. We evaluate our method on two public benchmark datasets for retinal disease diagnosis. The experimental results demonstrate that our method clearly outperforms other self-supervised feature learning methods and is comparable to the supervised baseline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge