Wenjian Qin
GraphMMP: A Graph Neural Network Model with Mutual Information and Global Fusion for Multimodal Medical Prognosis
Aug 24, 2025Abstract:In the field of multimodal medical data analysis, leveraging diverse types of data and understanding their hidden relationships continues to be a research focus. The main challenges lie in effectively modeling the complex interactions between heterogeneous data modalities with distinct characteristics while capturing both local and global dependencies across modalities. To address these challenges, this paper presents a two-stage multimodal prognosis model, GraphMMP, which is based on graph neural networks. The proposed model constructs feature graphs using mutual information and features a global fusion module built on Mamba, which significantly boosts prognosis performance. Empirical results show that GraphMMP surpasses existing methods on datasets related to liver prognosis and the METABRIC study, demonstrating its effectiveness in multimodal medical prognosis tasks.
Unpaired Multi-Domain Histopathology Virtual Staining using Dual Path Prompted Inversion
Dec 15, 2024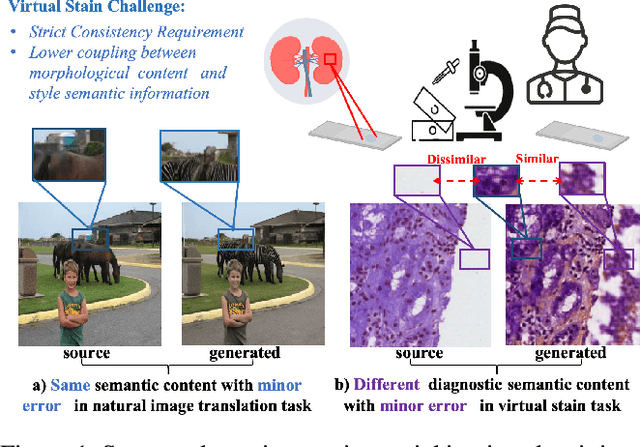


Abstract:Virtual staining leverages computer-aided techniques to transfer the style of histochemically stained tissue samples to other staining types. In virtual staining of pathological images, maintaining strict structural consistency is crucial, as these images emphasize structural integrity more than natural images. Even slight structural alterations can lead to deviations in diagnostic semantic information. Furthermore, the unpaired characteristic of virtual staining data may compromise the preservation of pathological diagnostic content. To address these challenges, we propose a dual-path inversion virtual staining method using prompt learning, which optimizes visual prompts to control content and style, while preserving complete pathological diagnostic content. Our proposed inversion technique comprises two key components: (1) Dual Path Prompted Strategy, we utilize a feature adapter function to generate reference images for inversion, providing style templates for input image inversion, called Style Target Path. We utilize the inversion of the input image as the Structural Target path, employing visual prompt images to maintain structural consistency in this path while preserving style information from the style Target path. During the deterministic sampling process, we achieve complete content-style disentanglement through a plug-and-play embedding visual prompt approach. (2) StainPrompt Optimization, where we only optimize the null visual prompt as ``operator'' for dual path inversion, rather than fine-tune pre-trained model. We optimize null visual prompt for structual and style trajectory around pivotal noise on each timestep, ensuring accurate dual-path inversion reconstruction. Extensive evaluations on publicly available multi-domain unpaired staining datasets demonstrate high structural consistency and accurate style transfer results.
Pathological Semantics-Preserving Learning for H&E-to-IHC Virtual Staining
Jul 04, 2024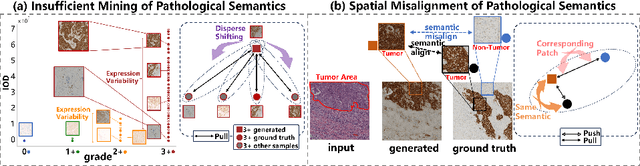
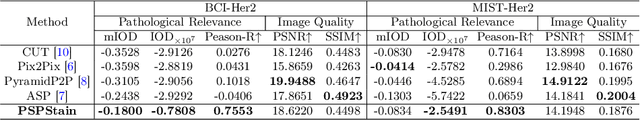
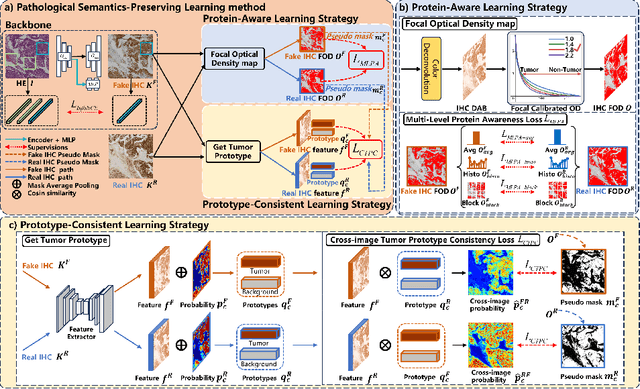
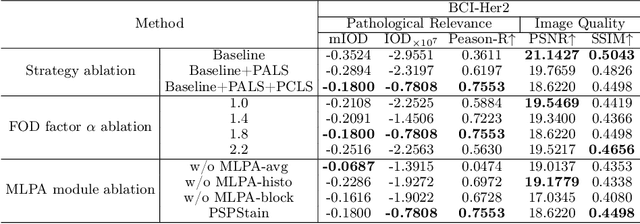
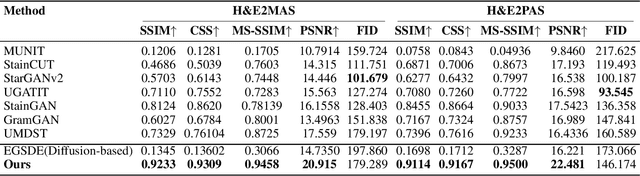
Abstract:Conventional hematoxylin-eosin (H&E) staining is limited to revealing cell morphology and distribution, whereas immunohistochemical (IHC) staining provides precise and specific visualization of protein activation at the molecular level. Virtual staining technology has emerged as a solution for highly efficient IHC examination, which directly transforms H&E-stained images to IHC-stained images. However, virtual staining is challenged by the insufficient mining of pathological semantics and the spatial misalignment of pathological semantics. To address these issues, we propose the Pathological Semantics-Preserving Learning method for Virtual Staining (PSPStain), which directly incorporates the molecular-level semantic information and enhances semantics interaction despite any spatial inconsistency. Specifically, PSPStain comprises two novel learning strategies: 1) Protein-Aware Learning Strategy (PALS) with Focal Optical Density (FOD) map maintains the coherence of protein expression level, which represents molecular-level semantic information; 2) Prototype-Consistent Learning Strategy (PCLS), which enhances cross-image semantic interaction by prototypical consistency learning. We evaluate PSPStain on two public datasets using five metrics: three clinically relevant metrics and two for image quality. Extensive experiments indicate that PSPStain outperforms current state-of-the-art H&E-to-IHC virtual staining methods and demonstrates a high pathological correlation between the staging of real and virtual stains.
BS-Diff: Effective Bone Suppression Using Conditional Diffusion Models from Chest X-Ray Images
Nov 26, 2023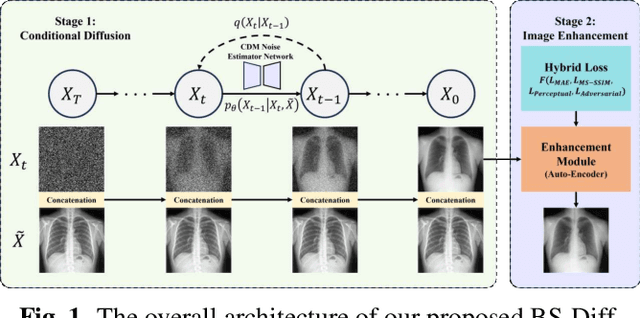
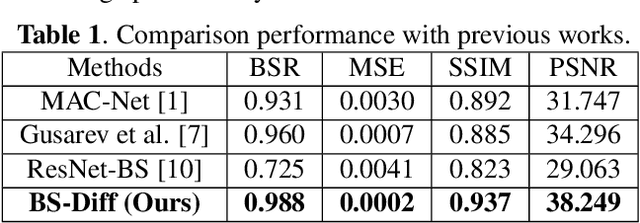
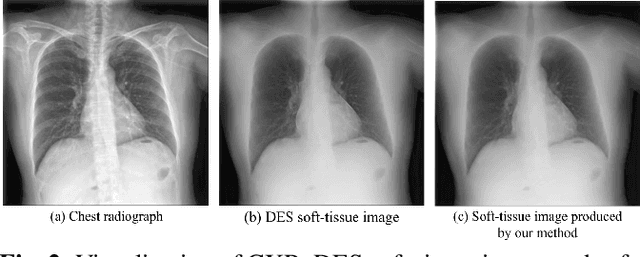
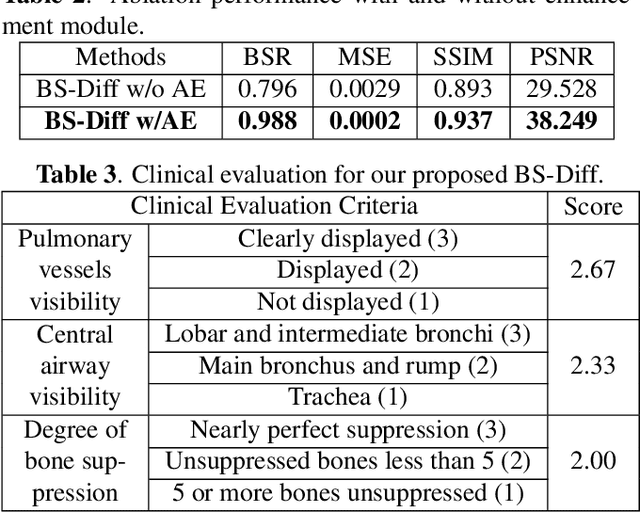
Abstract:Chest X-rays (CXRs) are commonly utilized as a low-dose modality for lung screening. Nonetheless, the efficacy of CXRs is somewhat impeded, given that approximately 75% of the lung area overlaps with bone, which in turn hampers the detection and diagnosis of diseases. As a remedial measure, bone suppression techniques have been introduced. The current dual-energy subtraction imaging technique in the clinic requires costly equipment and subjects being exposed to high radiation. To circumvent these issues, deep learning-based image generation algorithms have been proposed. However, existing methods fall short in terms of producing high-quality images and capturing texture details, particularly with pulmonary vessels. To address these issues, this paper proposes a new bone suppression framework, termed BS-Diff, that comprises a conditional diffusion model equipped with a U-Net architecture and a simple enhancement module to incorporate an autoencoder. Our proposed network cannot only generate soft tissue images with a high bone suppression rate but also possesses the capability to capture fine image details. Additionally, we compiled the largest dataset since 2010, including data from 120 patients with high-definition, high-resolution paired CXRs and soft tissue images collected by our affiliated hospital. Extensive experiments, comparative analyses, ablation studies, and clinical evaluations indicate that the proposed BS-Diff outperforms several bone-suppression models across multiple metrics.
A Hybrid Deep Feature-Based Deformable Image Registration Method for Pathological Images
Aug 17, 2022
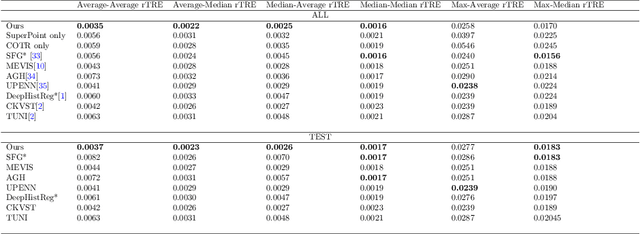
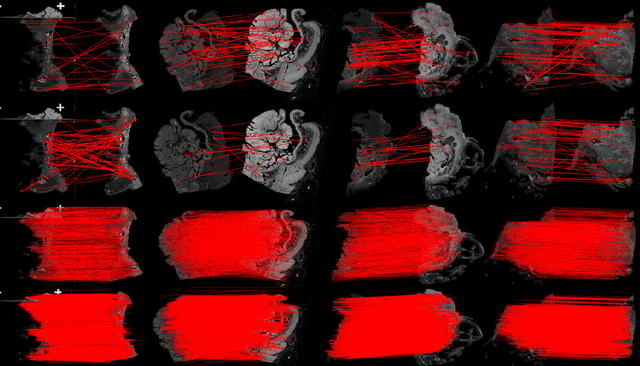
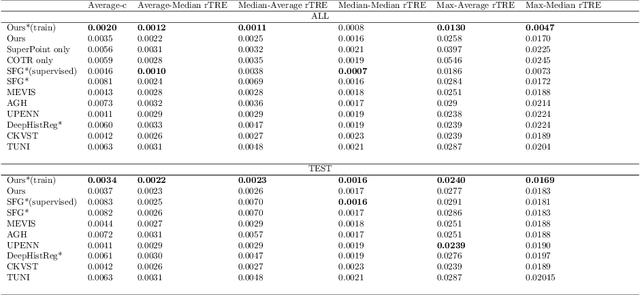
Abstract:Pathologists need to combine information from differently stained pathological slices to obtain accurate diagnostic results. Deformable image registration is a necessary technique for fusing multi-modal pathological slices. This paper proposes a hybrid deep feature-based deformable image registration framework for stained pathological samples. We first extract dense feature points and perform points matching by two deep learning feature networks. Then, to further reduce false matches, an outlier detection method combining the isolation forest statistical model and the local affine correction model is proposed. Finally, the interpolation method generates the DVF for pathology image registration based on the above matching points. We evaluate our method on the dataset of the Non-rigid Histology Image Registration (ANHIR) challenge, which is co-organized with the IEEE ISBI 2019 conference. Our technique outperforms the traditional approaches by 17% with the Average-Average registration target error (rTRE) reaching 0.0034. The proposed method achieved state-of-the-art performance and ranking it 1 in evaluating the test dataset. The proposed hybrid deep feature-based registration method can potentially become a reliable method for pathology image registration.
Total-Body Low-Dose CT Image Denoising using Prior Knowledge Transfer Technique with Contrastive Regularization Mechanism
Dec 06, 2021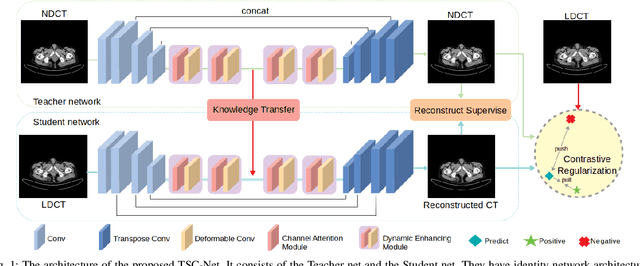
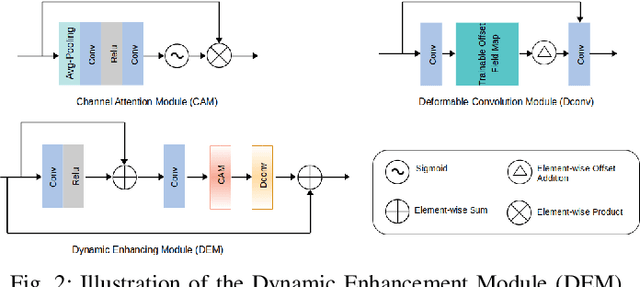
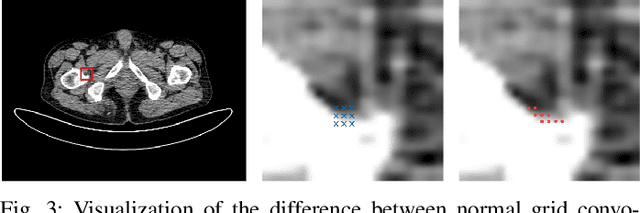
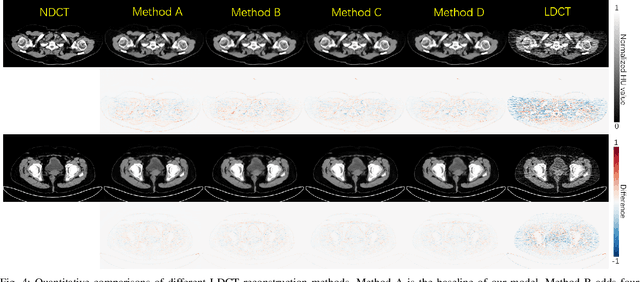
Abstract:Reducing the radiation exposure for patients in Total-body CT scans has attracted extensive attention in the medical imaging community. Given the fact that low radiation dose may result in increased noise and artifacts, which greatly affected the clinical diagnosis. To obtain high-quality Total-body Low-dose CT (LDCT) images, previous deep-learning-based research work has introduced various network architectures. However, most of these methods only adopt Normal-dose CT (NDCT) images as ground truths to guide the training of the denoising network. Such simple restriction leads the model to less effectiveness and makes the reconstructed images suffer from over-smoothing effects. In this paper, we propose a novel intra-task knowledge transfer method that leverages the distilled knowledge from NDCT images to assist the training process on LDCT images. The derived architecture is referred to as the Teacher-Student Consistency Network (TSC-Net), which consists of the teacher network and the student network with identical architecture. Through the supervision between intermediate features, the student network is encouraged to imitate the teacher network and gain abundant texture details. Moreover, to further exploit the information contained in CT scans, a contrastive regularization mechanism (CRM) built upon contrastive learning is introduced.CRM performs to pull the restored CT images closer to the NDCT samples and push far away from the LDCT samples in the latent space. In addition, based on the attention and deformable convolution mechanism, we design a Dynamic Enhancement Module (DEM) to improve the network transformation capability.
Elastic Net based Feature Ranking and Selection
Dec 30, 2020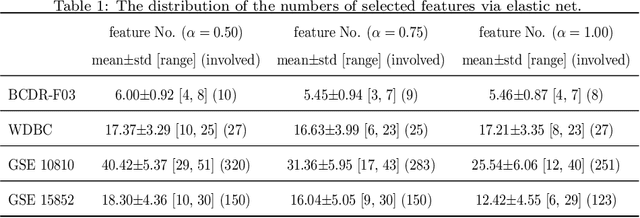
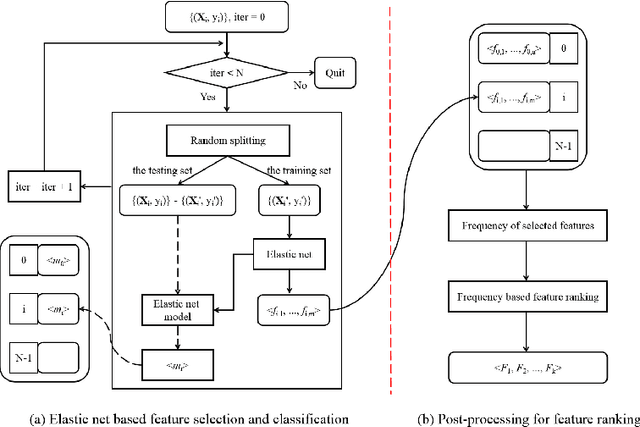
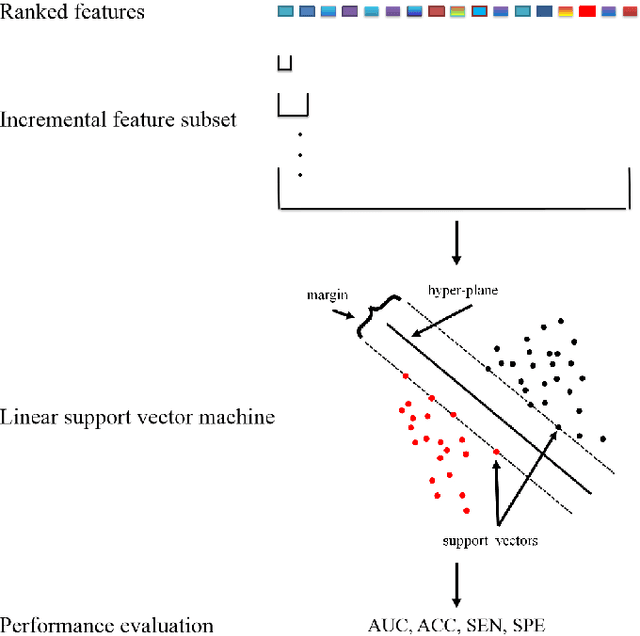
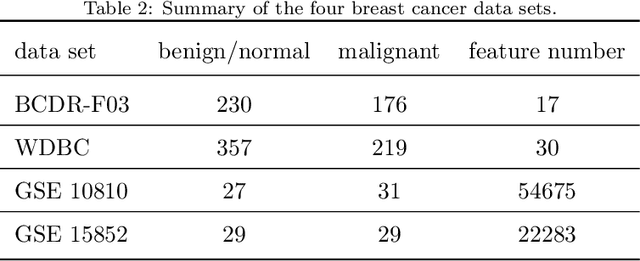
Abstract:Feature selection is important in data representation and intelligent diagnosis. Elastic net is one of the most widely used feature selectors. However, the features selected are dependant on the training data, and their weights dedicated for regularized regression are irrelevant to their importance if used for feature ranking, that degrades the model interpretability and extension. In this study, an intuitive idea is put at the end of multiple times of data splitting and elastic net based feature selection. It concerns the frequency of selected features and uses the frequency as an indicator of feature importance. After features are sorted according to their frequency, linear support vector machine performs the classification in an incremental manner. At last, a compact subset of discriminative features is selected by comparing the prediction performance. Experimental results on breast cancer data sets (BCDR-F03, WDBC, GSE 10810, and GSE 15852) suggest that the proposed framework achieves competitive or superior performance to elastic net and with consistent selection of fewer features. How to further enhance its consistency on high-dimension small-sample-size data sets should be paid more attention in our future work. The proposed framework is accessible online (https://github.com/NicoYuCN/elasticnetFR).
A Matlab Toolbox for Feature Importance Ranking
Mar 10, 2020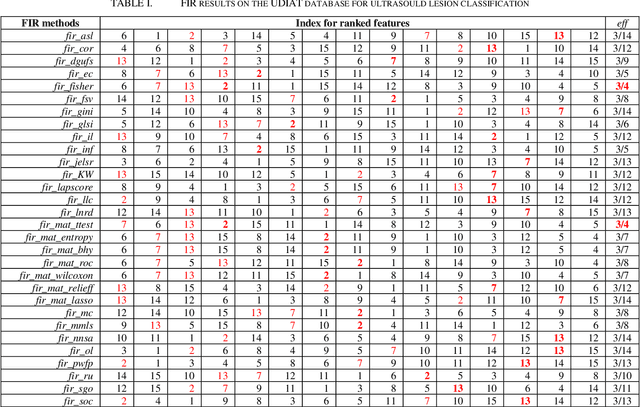
Abstract:More attention is being paid for feature importance ranking (FIR), in particular when thousands of features can be extracted for intelligent diagnosis and personalized medicine. A large number of FIR approaches have been proposed, while few are integrated for comparison and real-life applications. In this study, a matlab toolbox is presented and a total of 30 algorithms are collected. Moreover, the toolbox is evaluated on a database of 163 ultrasound images. To each breast mass lesion, 15 features are extracted. To figure out the optimal subset of features for classification, all combinations of features are tested and linear support vector machine is used for the malignancy prediction of lesions annotated in ultrasound images. At last, the effectiveness of FIR is analyzed according to performance comparison. The toolbox is online (https://github.com/NicoYuCN/matFIR). In our future work, more FIR methods, feature selection methods and machine learning classifiers will be integrated.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge