Jingjing Dai
Three-Dimensional Medical Image Fusion with Deformable Cross-Attention
Oct 10, 2023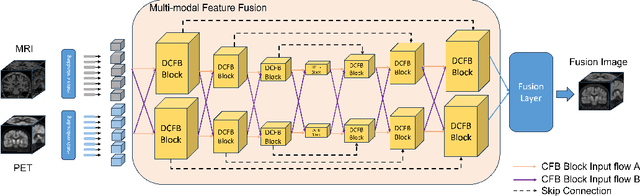
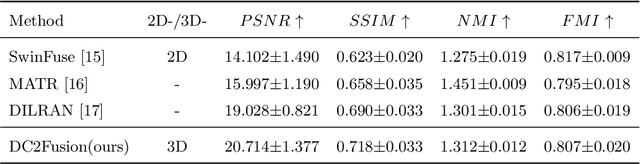
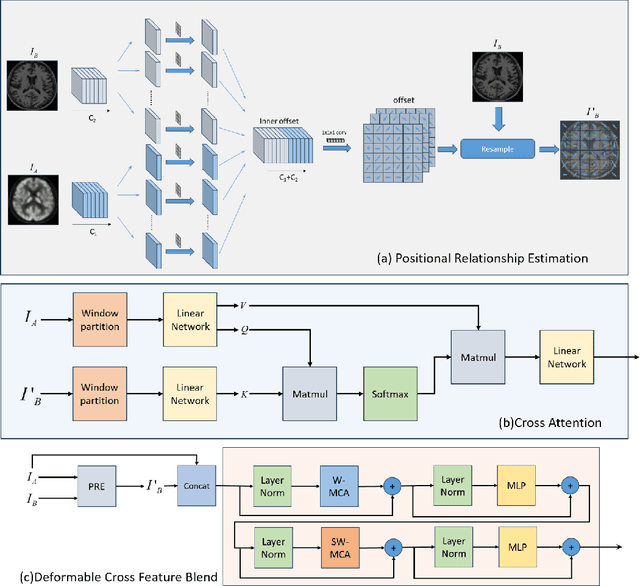
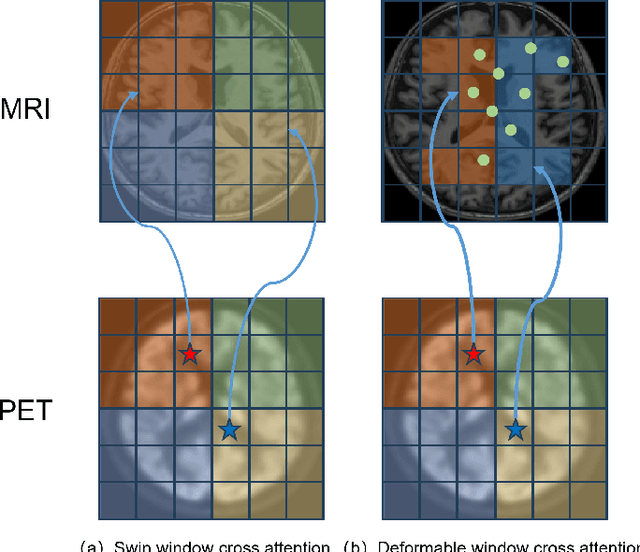
Abstract:Multimodal medical image fusion plays an instrumental role in several areas of medical image processing, particularly in disease recognition and tumor detection. Traditional fusion methods tend to process each modality independently before combining the features and reconstructing the fusion image. However, this approach often neglects the fundamental commonalities and disparities between multimodal information. Furthermore, the prevailing methodologies are largely confined to fusing two-dimensional (2D) medical image slices, leading to a lack of contextual supervision in the fusion images and subsequently, a decreased information yield for physicians relative to three-dimensional (3D) images. In this study, we introduce an innovative unsupervised feature mutual learning fusion network designed to rectify these limitations. Our approach incorporates a Deformable Cross Feature Blend (DCFB) module that facilitates the dual modalities in discerning their respective similarities and differences. We have applied our model to the fusion of 3D MRI and PET images obtained from 660 patients in the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset. Through the application of the DCFB module, our network generates high-quality MRI-PET fusion images. Experimental results demonstrate that our method surpasses traditional 2D image fusion methods in performance metrics such as Peak Signal to Noise Ratio (PSNR) and Structural Similarity Index Measure (SSIM). Importantly, the capacity of our method to fuse 3D images enhances the information available to physicians and researchers, thus marking a significant step forward in the field. The code will soon be available online.
QUIZ: An Arbitrary Volumetric Point Matching Method for Medical Image Registration
Sep 30, 2023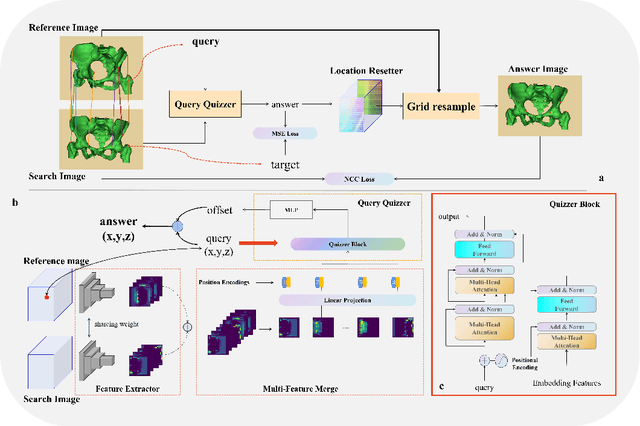
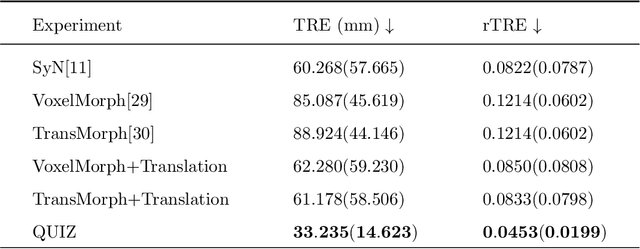
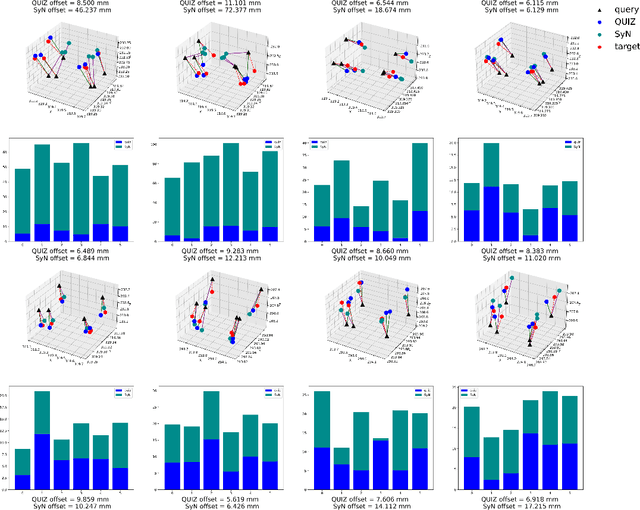
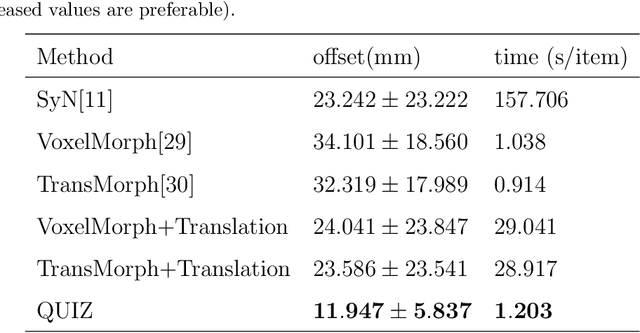
Abstract:Rigid pre-registration involving local-global matching or other large deformation scenarios is crucial. Current popular methods rely on unsupervised learning based on grayscale similarity, but under circumstances where different poses lead to varying tissue structures, or where image quality is poor, these methods tend to exhibit instability and inaccuracies. In this study, we propose a novel method for medical image registration based on arbitrary voxel point of interest matching, called query point quizzer (QUIZ). QUIZ focuses on the correspondence between local-global matching points, specifically employing CNN for feature extraction and utilizing the Transformer architecture for global point matching queries, followed by applying average displacement for local image rigid transformation. We have validated this approach on a large deformation dataset of cervical cancer patients, with results indicating substantially smaller deviations compared to state-of-the-art methods. Remarkably, even for cross-modality subjects, it achieves results surpassing the current state-of-the-art.
XTransCT: Ultra-Fast Volumetric CT Reconstruction using Two Orthogonal X-Ray Projections via a Transformer Network
May 31, 2023Abstract:Computed tomography (CT) scans offer a detailed, three-dimensional representation of patients' internal organs. However, conventional CT reconstruction techniques necessitate acquiring hundreds or thousands of x-ray projections through a complete rotational scan of the body, making navigation or positioning during surgery infeasible. In image-guided radiation therapy, a method that reconstructs ultra-sparse X-ray projections into CT images, we can exploit the substantially reduced radiation dose and minimize equipment burden for localization and navigation. In this study, we introduce a novel Transformer architecture, termed XTransCT, devised to facilitate real-time reconstruction of CT images from two-dimensional X-ray images. We assess our approach regarding image quality and structural reliability using a dataset of fifty patients, supplied by a hospital, as well as the larger public dataset LIDC-IDRI, which encompasses thousands of patients. Additionally, we validated our algorithm's generalizability on the LNDb dataset. Our findings indicate that our algorithm surpasses other methods in image quality, structural precision, and generalizability. Moreover, in comparison to previous 3D convolution-based approaches, we note a substantial speed increase of approximately 300 $\%$, achieving 44 ms per 3D image reconstruction. To guarantee the replicability of our results, we have made our code publicly available.
A Hybrid Deep Feature-Based Deformable Image Registration Method for Pathological Images
Aug 17, 2022
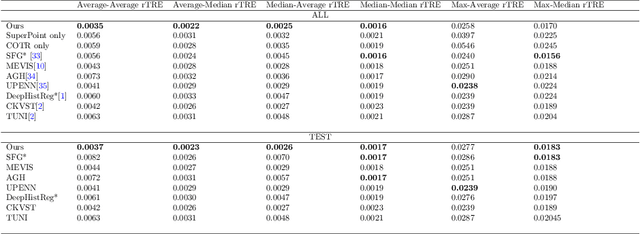
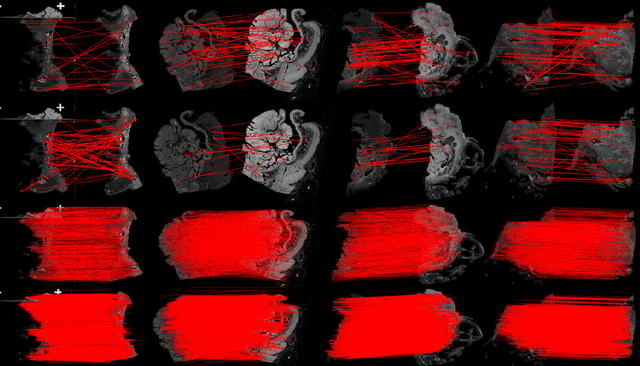
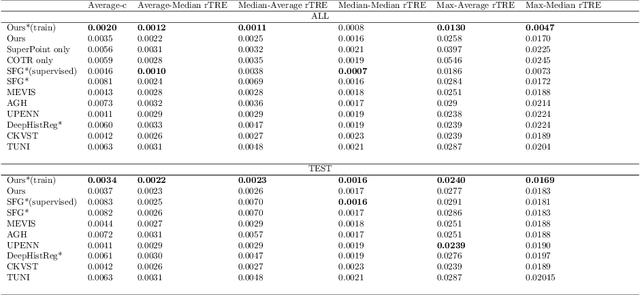
Abstract:Pathologists need to combine information from differently stained pathological slices to obtain accurate diagnostic results. Deformable image registration is a necessary technique for fusing multi-modal pathological slices. This paper proposes a hybrid deep feature-based deformable image registration framework for stained pathological samples. We first extract dense feature points and perform points matching by two deep learning feature networks. Then, to further reduce false matches, an outlier detection method combining the isolation forest statistical model and the local affine correction model is proposed. Finally, the interpolation method generates the DVF for pathology image registration based on the above matching points. We evaluate our method on the dataset of the Non-rigid Histology Image Registration (ANHIR) challenge, which is co-organized with the IEEE ISBI 2019 conference. Our technique outperforms the traditional approaches by 17% with the Average-Average registration target error (rTRE) reaching 0.0034. The proposed method achieved state-of-the-art performance and ranking it 1 in evaluating the test dataset. The proposed hybrid deep feature-based registration method can potentially become a reliable method for pathology image registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge