Xinxin Fan
SifterNet: A Generalized and Model-Agnostic Trigger Purification Approach
May 20, 2025Abstract:Aiming at resisting backdoor attacks in convolution neural networks and vision Transformer-based large model, this paper proposes a generalized and model-agnostic trigger-purification approach resorting to the classic Ising model. To date, existing trigger detection/removal studies usually require to know the detailed knowledge of target model in advance, access to a large number of clean samples or even model-retraining authorization, which brings the huge inconvenience for practical applications, especially inaccessible to target model. An ideal countermeasure ought to eliminate the implanted trigger without regarding whatever the target models are. To this end, a lightweight and black-box defense approach SifterNet is proposed through leveraging the memorization-association functionality of Hopfield network, by which the triggers of input samples can be effectively purified in a proper manner. The main novelty of our proposed approach lies in the introduction of ideology of Ising model. Extensive experiments also validate the effectiveness of our approach in terms of proper trigger purification and high accuracy achievement, and compared to the state-of-the-art baselines under several commonly-used datasets, our SiferNet has a significant superior performance.
Adverseness vs. Equilibrium: Exploring Graph Adversarial Resilience through Dynamic Equilibrium
May 20, 2025Abstract:Adversarial attacks to graph analytics are gaining increased attention. To date, two lines of countermeasures have been proposed to resist various graph adversarial attacks from the perspectives of either graph per se or graph neural networks. Nevertheless, a fundamental question lies in whether there exists an intrinsic adversarial resilience state within a graph regime and how to find out such a critical state if exists. This paper contributes to tackle the above research questions from three unique perspectives: i) we regard the process of adversarial learning on graph as a complex multi-object dynamic system, and model the behavior of adversarial attack; ii) we propose a generalized theoretical framework to show the existence of critical adversarial resilience state; and iii) we develop a condensed one-dimensional function to capture the dynamic variation of graph regime under perturbations, and pinpoint the critical state through solving the equilibrium point of dynamic system. Multi-facet experiments are conducted to show our proposed approach can significantly outperform the state-of-the-art defense methods under five commonly-used real-world datasets and three representative attacks.
Multimodal Information Interaction for Medical Image Segmentation
Apr 25, 2024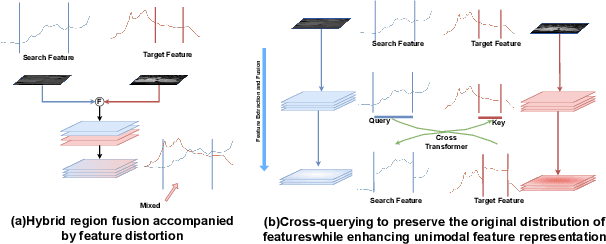
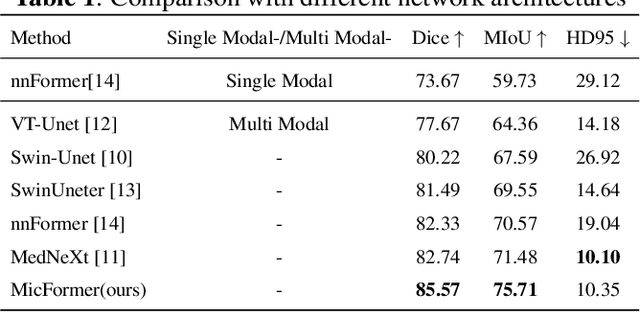
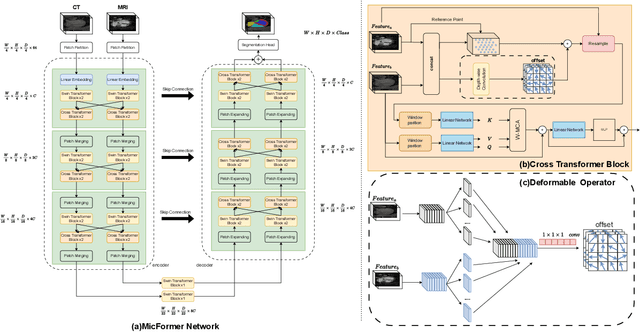
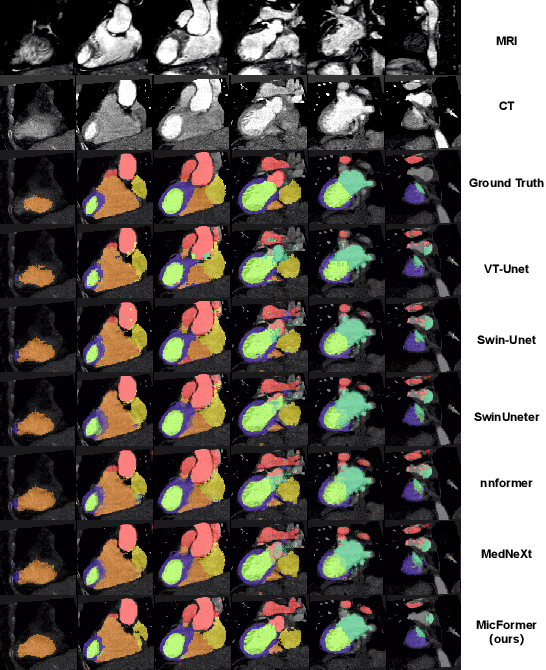
Abstract:The use of multimodal data in assisted diagnosis and segmentation has emerged as a prominent area of interest in current research. However, one of the primary challenges is how to effectively fuse multimodal features. Most of the current approaches focus on the integration of multimodal features while ignoring the correlation and consistency between different modal features, leading to the inclusion of potentially irrelevant information. To address this issue, we introduce an innovative Multimodal Information Cross Transformer (MicFormer), which employs a dual-stream architecture to simultaneously extract features from each modality. Leveraging the Cross Transformer, it queries features from one modality and retrieves corresponding responses from another, facilitating effective communication between bimodal features. Additionally, we incorporate a deformable Transformer architecture to expand the search space. We conducted experiments on the MM-WHS dataset, and in the CT-MRI multimodal image segmentation task, we successfully improved the whole-heart segmentation DICE score to 85.57 and MIoU to 75.51. Compared to other multimodal segmentation techniques, our method outperforms by margins of 2.83 and 4.23, respectively. This demonstrates the efficacy of MicFormer in integrating relevant information between different modalities in multimodal tasks. These findings hold significant implications for multimodal image tasks, and we believe that MicFormer possesses extensive potential for broader applications across various domains. Access to our method is available at https://github.com/fxxJuses/MICFormer
Contrastive Prompt Learning-based Code Search based on Interaction Matrix
Oct 10, 2023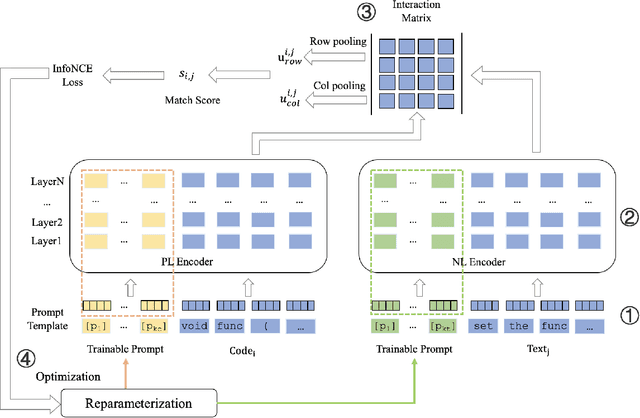
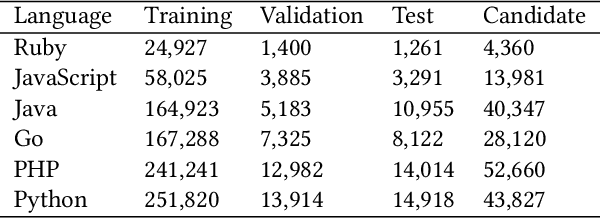
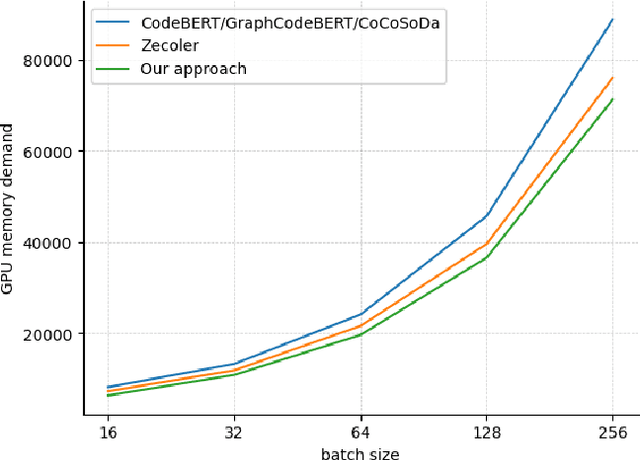
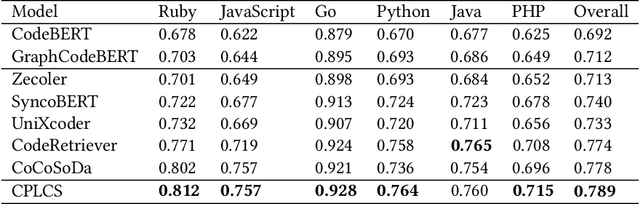
Abstract:Code search aims to retrieve the code snippet that highly matches the given query described in natural language. Recently, many code pre-training approaches have demonstrated impressive performance on code search. However, existing code search methods still suffer from two performance constraints: inadequate semantic representation and the semantic gap between natural language (NL) and programming language (PL). In this paper, we propose CPLCS, a contrastive prompt learning-based code search method based on the cross-modal interaction mechanism. CPLCS comprises:(1) PL-NL contrastive learning, which learns the semantic matching relationship between PL and NL representations; (2) a prompt learning design for a dual-encoder structure that can alleviate the problem of inadequate semantic representation; (3) a cross-modal interaction mechanism to enhance the fine-grained mapping between NL and PL. We conduct extensive experiments to evaluate the effectiveness of our approach on a real-world dataset across six programming languages. The experiment results demonstrate the efficacy of our approach in improving semantic representation quality and mapping ability between PL and NL.
Three-Dimensional Medical Image Fusion with Deformable Cross-Attention
Oct 10, 2023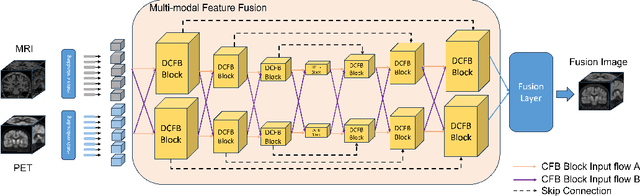
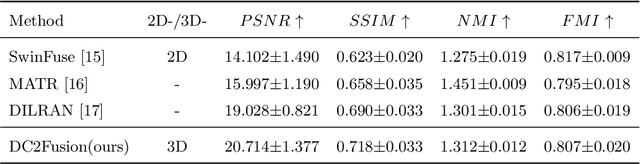
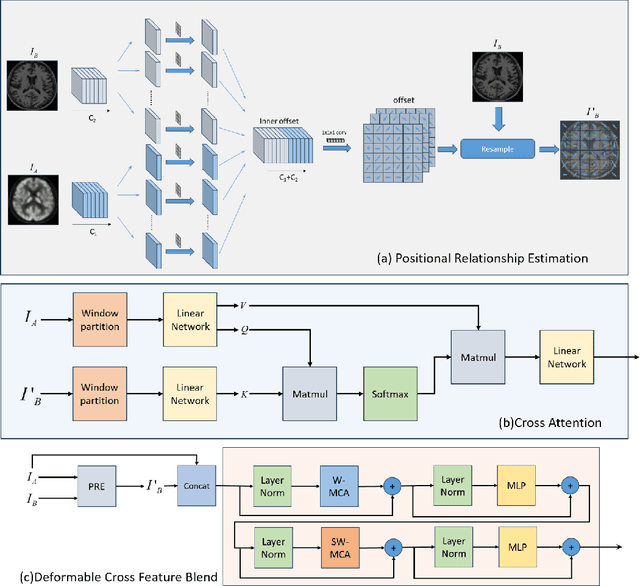
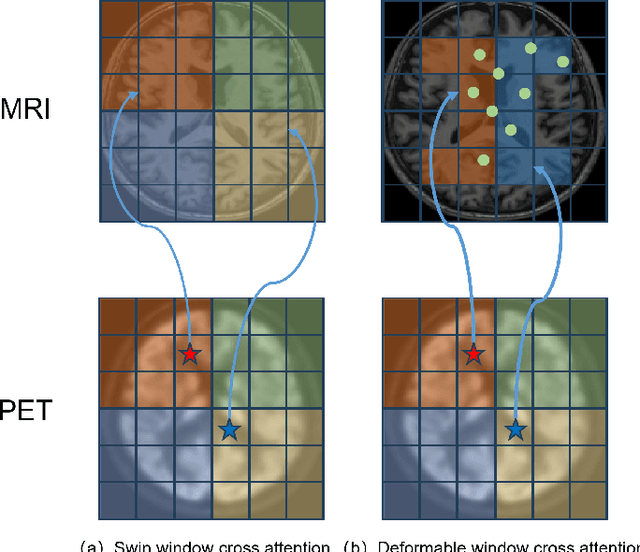
Abstract:Multimodal medical image fusion plays an instrumental role in several areas of medical image processing, particularly in disease recognition and tumor detection. Traditional fusion methods tend to process each modality independently before combining the features and reconstructing the fusion image. However, this approach often neglects the fundamental commonalities and disparities between multimodal information. Furthermore, the prevailing methodologies are largely confined to fusing two-dimensional (2D) medical image slices, leading to a lack of contextual supervision in the fusion images and subsequently, a decreased information yield for physicians relative to three-dimensional (3D) images. In this study, we introduce an innovative unsupervised feature mutual learning fusion network designed to rectify these limitations. Our approach incorporates a Deformable Cross Feature Blend (DCFB) module that facilitates the dual modalities in discerning their respective similarities and differences. We have applied our model to the fusion of 3D MRI and PET images obtained from 660 patients in the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset. Through the application of the DCFB module, our network generates high-quality MRI-PET fusion images. Experimental results demonstrate that our method surpasses traditional 2D image fusion methods in performance metrics such as Peak Signal to Noise Ratio (PSNR) and Structural Similarity Index Measure (SSIM). Importantly, the capacity of our method to fuse 3D images enhances the information available to physicians and researchers, thus marking a significant step forward in the field. The code will soon be available online.
QUIZ: An Arbitrary Volumetric Point Matching Method for Medical Image Registration
Sep 30, 2023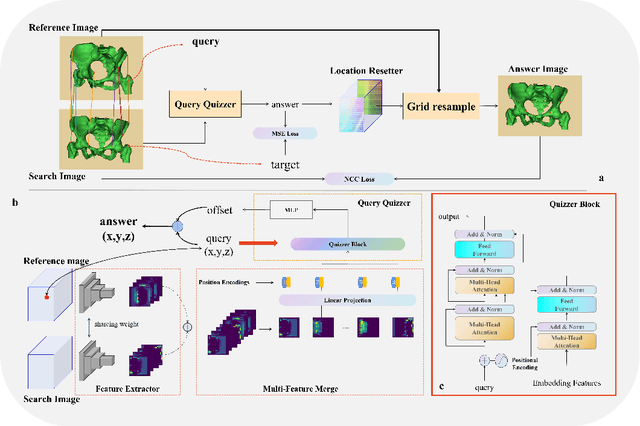
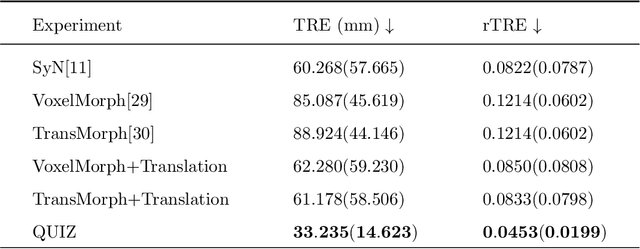
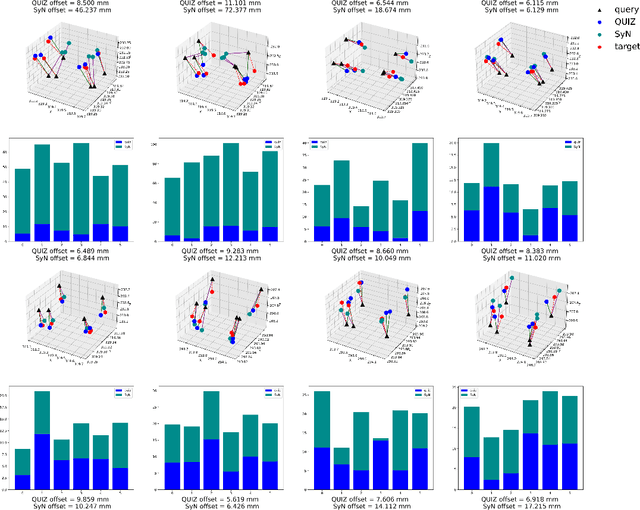
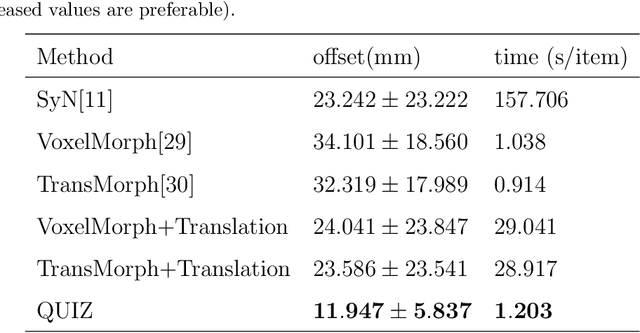
Abstract:Rigid pre-registration involving local-global matching or other large deformation scenarios is crucial. Current popular methods rely on unsupervised learning based on grayscale similarity, but under circumstances where different poses lead to varying tissue structures, or where image quality is poor, these methods tend to exhibit instability and inaccuracies. In this study, we propose a novel method for medical image registration based on arbitrary voxel point of interest matching, called query point quizzer (QUIZ). QUIZ focuses on the correspondence between local-global matching points, specifically employing CNN for feature extraction and utilizing the Transformer architecture for global point matching queries, followed by applying average displacement for local image rigid transformation. We have validated this approach on a large deformation dataset of cervical cancer patients, with results indicating substantially smaller deviations compared to state-of-the-art methods. Remarkably, even for cross-modality subjects, it achieves results surpassing the current state-of-the-art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge