Wenfeng He
XTransCT: Ultra-Fast Volumetric CT Reconstruction using Two Orthogonal X-Ray Projections via a Transformer Network
May 31, 2023Abstract:Computed tomography (CT) scans offer a detailed, three-dimensional representation of patients' internal organs. However, conventional CT reconstruction techniques necessitate acquiring hundreds or thousands of x-ray projections through a complete rotational scan of the body, making navigation or positioning during surgery infeasible. In image-guided radiation therapy, a method that reconstructs ultra-sparse X-ray projections into CT images, we can exploit the substantially reduced radiation dose and minimize equipment burden for localization and navigation. In this study, we introduce a novel Transformer architecture, termed XTransCT, devised to facilitate real-time reconstruction of CT images from two-dimensional X-ray images. We assess our approach regarding image quality and structural reliability using a dataset of fifty patients, supplied by a hospital, as well as the larger public dataset LIDC-IDRI, which encompasses thousands of patients. Additionally, we validated our algorithm's generalizability on the LNDb dataset. Our findings indicate that our algorithm surpasses other methods in image quality, structural precision, and generalizability. Moreover, in comparison to previous 3D convolution-based approaches, we note a substantial speed increase of approximately 300 $\%$, achieving 44 ms per 3D image reconstruction. To guarantee the replicability of our results, we have made our code publicly available.
A Hybrid Deep Feature-Based Deformable Image Registration Method for Pathological Images
Aug 17, 2022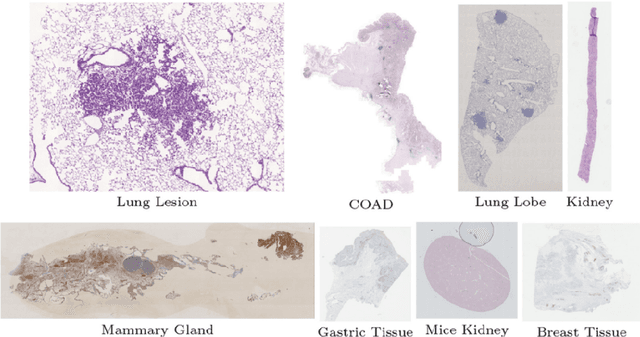
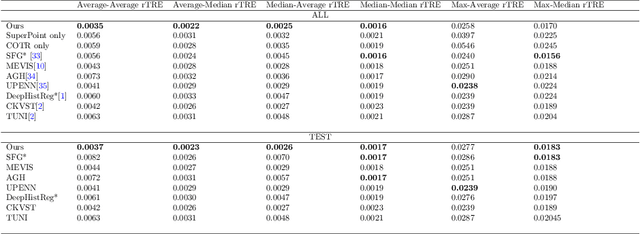
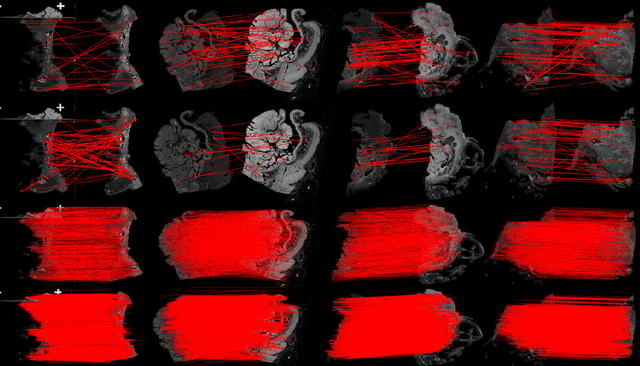
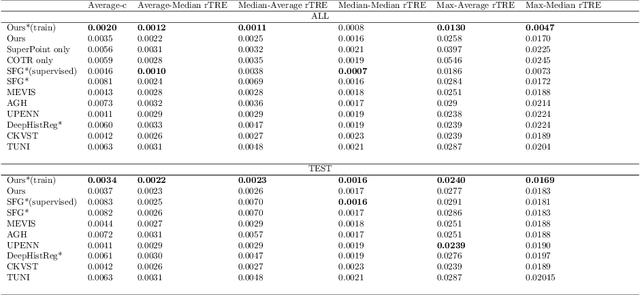
Abstract:Pathologists need to combine information from differently stained pathological slices to obtain accurate diagnostic results. Deformable image registration is a necessary technique for fusing multi-modal pathological slices. This paper proposes a hybrid deep feature-based deformable image registration framework for stained pathological samples. We first extract dense feature points and perform points matching by two deep learning feature networks. Then, to further reduce false matches, an outlier detection method combining the isolation forest statistical model and the local affine correction model is proposed. Finally, the interpolation method generates the DVF for pathology image registration based on the above matching points. We evaluate our method on the dataset of the Non-rigid Histology Image Registration (ANHIR) challenge, which is co-organized with the IEEE ISBI 2019 conference. Our technique outperforms the traditional approaches by 17% with the Average-Average registration target error (rTRE) reaching 0.0034. The proposed method achieved state-of-the-art performance and ranking it 1 in evaluating the test dataset. The proposed hybrid deep feature-based registration method can potentially become a reliable method for pathology image registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge