Brett K. Beaulieu-Jones
Harvard Medical School
Efficient Variance-reduced Estimation from Generative EHR Models: The SCOPE and REACH Estimators
Feb 03, 2026Abstract:Generative models trained using self-supervision of tokenized electronic health record (EHR) timelines show promise for clinical outcome prediction. This is typically done using Monte Carlo simulation for future patient trajectories. However, existing approaches suffer from three key limitations: sparse estimate distributions that poorly differentiate patient risk levels, extreme computational costs, and high sampling variance. We propose two new estimators: the Sum of Conditional Outcome Probability Estimator (SCOPE) and Risk Estimation from Anticipated Conditional Hazards (REACH), that leverage next-token probability distributions discarded by standard Monte Carlo. We prove both estimators are unbiased and that REACH guarantees variance reduction over Monte Carlo sampling for any model and outcome. Empirically, on hospital mortality prediction in MIMIC-IV using the ETHOS-ARES framework, SCOPE and REACH match 100-sample Monte Carlo performance using only 10-11 samples (95% CI: [9,11]), representing a ~10x reduction in inference cost without degrading calibration. For ICU admission prediction, efficiency gains are more modest (~1.2x), which we attribute to the outcome's lower "spontaneity," a property we characterize theoretically and empirically. These methods substantially improve the feasibility of deploying generative EHR models in resource-constrained clinical settings.
Quantifying surprise in clinical care: Detecting highly informative events in electronic health records with foundation models
Jul 30, 2025Abstract:We present a foundation model-derived method to identify highly informative tokens and events in electronic health records. Our approach considers incoming data in the entire context of a patient's hospitalization and so can flag anomalous events that rule-based approaches would consider within a normal range. We demonstrate that the events our model flags are significant for predicting downstream patient outcomes and that a fraction of events identified as carrying little information can safely be dropped. Additionally, we show how informativeness can help interpret the predictions of prognostic models trained on foundation model-derived representations.
Foundation models for electronic health records: representation dynamics and transferability
Apr 14, 2025Abstract:Foundation models (FMs) trained on electronic health records (EHRs) have shown strong performance on a range of clinical prediction tasks. However, adapting these models to local health systems remains challenging due to limited data availability and resource constraints. In this study, we investigated what these models learn and evaluated the transferability of an FM trained on MIMIC-IV to an institutional EHR dataset at the University of Chicago Medical Center. We assessed their ability to identify outlier patients and examined representation-space patient trajectories in relation to future clinical outcomes. We also evaluated the performance of supervised fine-tuned classifiers on both source and target datasets. Our findings offer insights into the adaptability of FMs across different healthcare systems, highlight considerations for their effective implementation, and provide an empirical analysis of the underlying factors that contribute to their predictive performance.
ML4H Abstract Track 2019
Feb 05, 2020Abstract:A collection of the accepted abstracts for the Machine Learning for Health (ML4H) workshop at NeurIPS 2019. This index is not complete, as some accepted abstracts chose to opt-out of inclusion.
Privacy-Preserving Distributed Deep Learning for Clinical Data
Dec 04, 2018Abstract:Deep learning with medical data often requires larger samples sizes than are available at single providers. While data sharing among institutions is desirable to train more accurate and sophisticated models, it can lead to severe privacy concerns due the sensitive nature of the data. This problem has motivated a number of studies on distributed training of neural networks that do not require direct sharing of the training data. However, simple distributed training does not offer provable privacy guarantees to satisfy technical safe standards and may reveal information about the underlying patients. We present a method to train neural networks for clinical data in a distributed fashion under differential privacy. We demonstrate these methods on two datasets that include information from multiple independent sites, the eICU collaborative Research Database and The Cancer Genome Atlas.
Machine Learning for Health (ML4H) Workshop at NeurIPS 2018
Nov 24, 2018Abstract:This volume represents the accepted submissions from the Machine Learning for Health (ML4H) workshop at the conference on Neural Information Processing Systems (NeurIPS) 2018, held on December 8, 2018 in Montreal, Canada.
Learning Contextual Hierarchical Structure of Medical Concepts with Poincairé Embeddings to Clarify Phenotypes
Nov 03, 2018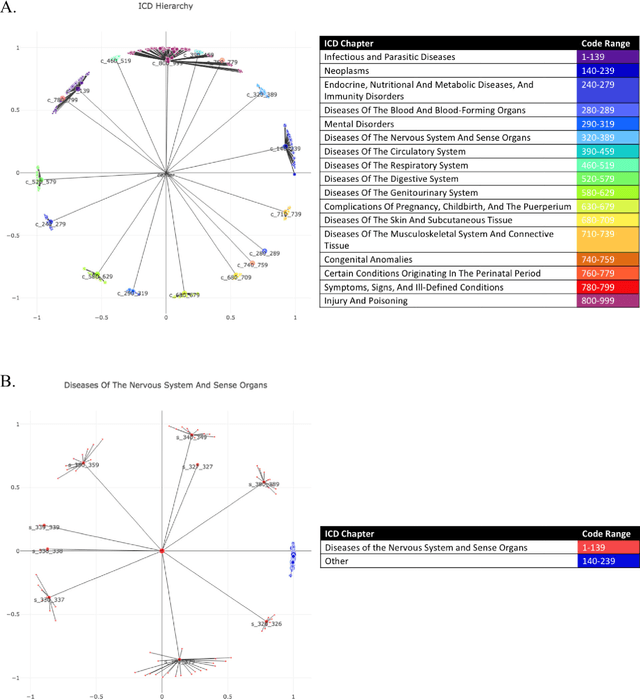
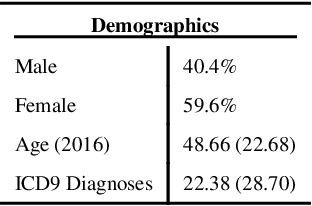
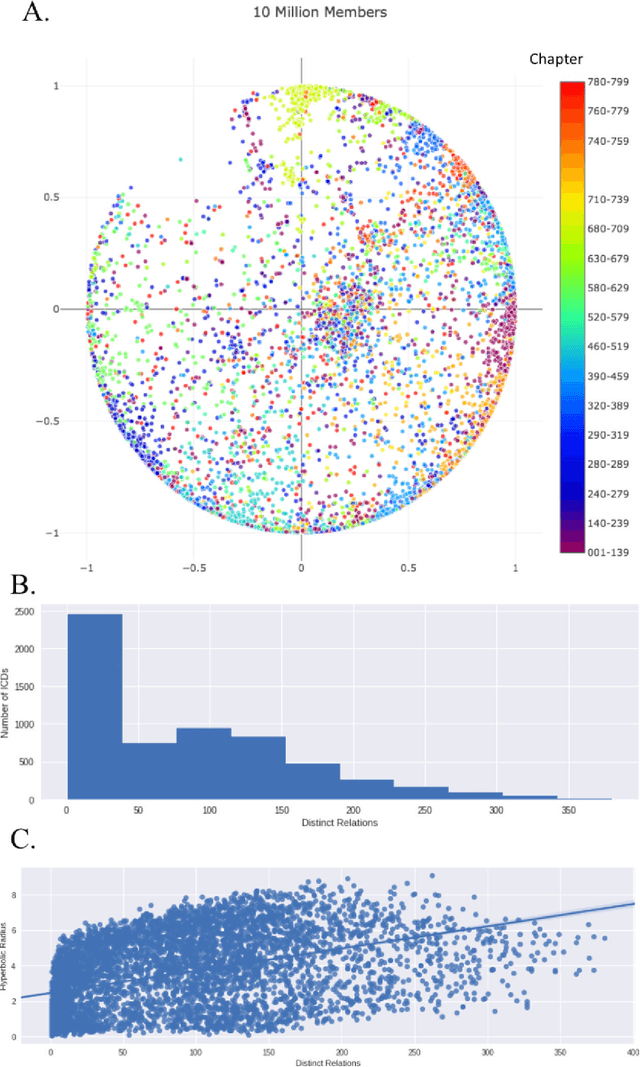
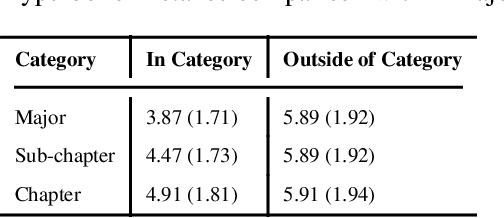
Abstract:Biomedical association studies are increasingly done using clinical concepts, and in particular diagnostic codes from clinical data repositories as phenotypes. Clinical concepts can be represented in a meaningful, vector space using word embedding models. These embeddings allow for comparison between clinical concepts or for straightforward input to machine learning models. Using traditional approaches, good representations require high dimensionality, making downstream tasks such as visualization more difficult. We applied Poincar\'e embeddings in a 2-dimensional hyperbolic space to a large-scale administrative claims database and show performance comparable to 100-dimensional embeddings in a euclidean space. We then examine disease relationships under different disease contexts to better understand potential phenotypes.
Machine Learning for Structured Clinical Data
Jul 21, 2017


Abstract:Research is a tertiary priority in the EHR, where the priorities are patient care and billing. Because of this, the data is not standardized or formatted in a manner easily adapted to machine learning approaches. Data may be missing for a large variety of reasons ranging from individual input styles to differences in clinical decision making, for example, which lab tests to issue. Few patients are annotated at a research quality, limiting sample size and presenting a moving gold standard. Patient progression over time is key to understanding many diseases but many machine learning algorithms require a snapshot, at a single time point, to create a usable vector form. Furthermore, algorithms that produce black box results do not provide the interpretability required for clinical adoption. This chapter discusses these challenges and others in applying machine learning techniques to the structured EHR (i.e. Patient Demographics, Family History, Medication Information, Vital Signs, Laboratory Tests, Genetic Testing). It does not cover feature extraction from additional sources such as imaging data or free text patient notes but the approaches discussed can include features extracted from these sources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge