Learning Contextual Hierarchical Structure of Medical Concepts with Poincairé Embeddings to Clarify Phenotypes
Paper and Code
Nov 03, 2018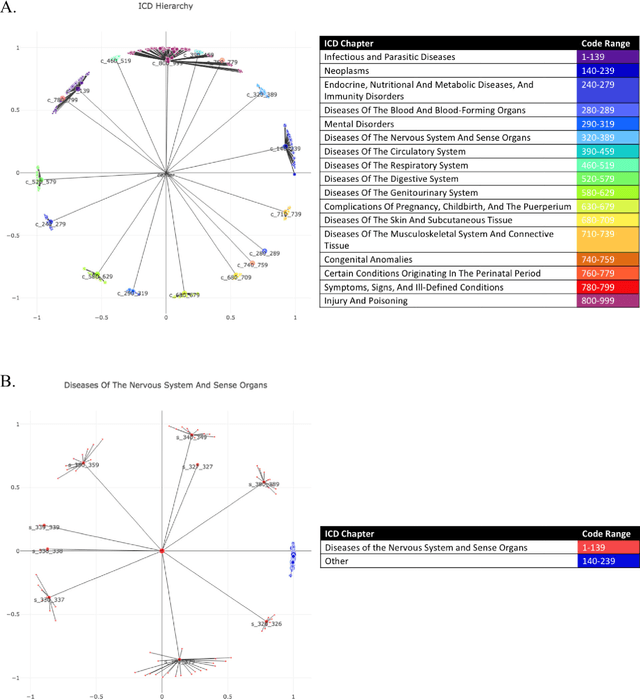
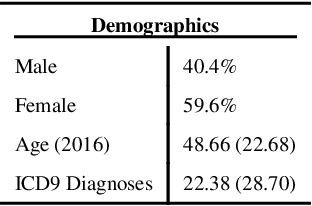
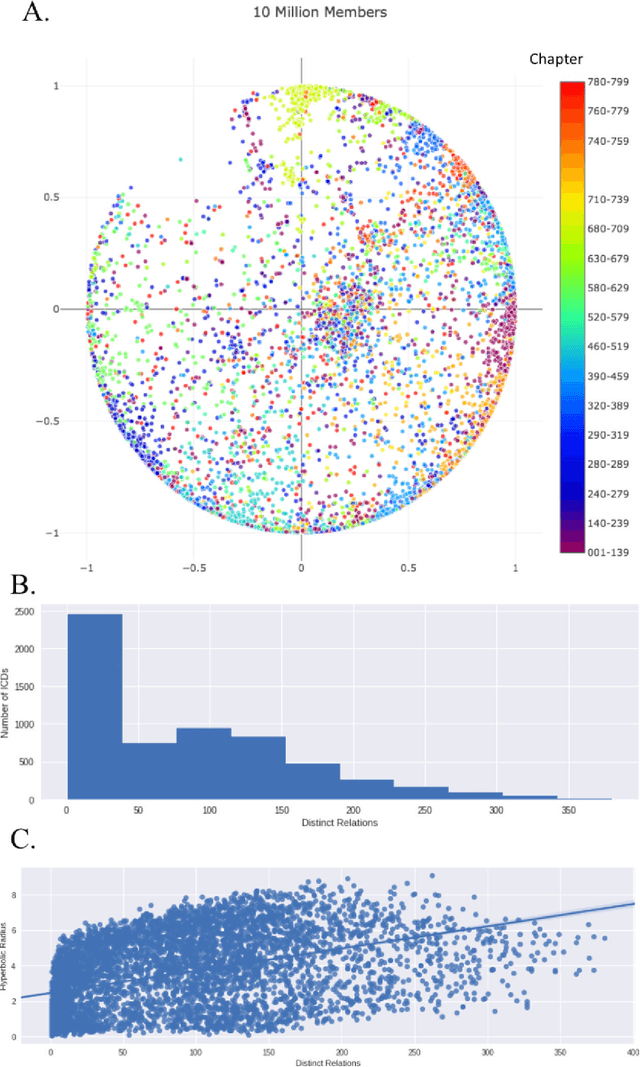
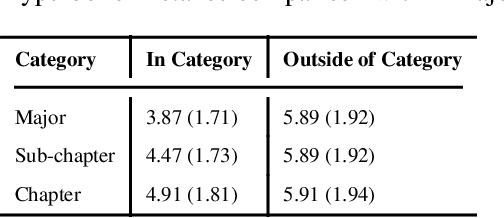
Biomedical association studies are increasingly done using clinical concepts, and in particular diagnostic codes from clinical data repositories as phenotypes. Clinical concepts can be represented in a meaningful, vector space using word embedding models. These embeddings allow for comparison between clinical concepts or for straightforward input to machine learning models. Using traditional approaches, good representations require high dimensionality, making downstream tasks such as visualization more difficult. We applied Poincar\'e embeddings in a 2-dimensional hyperbolic space to a large-scale administrative claims database and show performance comparable to 100-dimensional embeddings in a euclidean space. We then examine disease relationships under different disease contexts to better understand potential phenotypes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge