Inbar Fried
Safe Start Regions for Medical Steerable Needle Automation
Apr 12, 2024Abstract:Steerable needles are minimally invasive devices that enable novel medical procedures by following curved paths to avoid critical anatomical obstacles. Planning algorithms can be used to find a steerable needle motion plan to a target. Deployment typically consists of a physician manually inserting the steerable needle into tissue at the motion plan's start pose and handing off control to a robot, which then autonomously steers it to the target along the plan. The handoff between human and robot is critical for procedure success, as even small deviations from the start pose change the steerable needle's workspace and there is no guarantee that the target will still be reachable. We introduce a metric that evaluates the robustness to such start pose deviations. When measuring this robustness to deviations, we consider the tradeoff between being robust to changes in position versus changes in orientation. We evaluate our metric through simulation in an abstract, a liver, and a lung planning scenario. Our evaluation shows that our metric can be combined with different motion planners and that it efficiently determines large, safe start regions.
Leveraging Near-Field Lighting for Monocular Depth Estimation from Endoscopy Videos
Mar 26, 2024



Abstract:Monocular depth estimation in endoscopy videos can enable assistive and robotic surgery to obtain better coverage of the organ and detection of various health issues. Despite promising progress on mainstream, natural image depth estimation, techniques perform poorly on endoscopy images due to a lack of strong geometric features and challenging illumination effects. In this paper, we utilize the photometric cues, i.e., the light emitted from an endoscope and reflected by the surface, to improve monocular depth estimation. We first create two novel loss functions with supervised and self-supervised variants that utilize a per-pixel shading representation. We then propose a novel depth refinement network (PPSNet) that leverages the same per-pixel shading representation. Finally, we introduce teacher-student transfer learning to produce better depth maps from both synthetic data with supervision and clinical data with self-supervision. We achieve state-of-the-art results on the C3VD dataset while estimating high-quality depth maps from clinical data. Our code, pre-trained models, and supplementary materials can be found on our project page: https://ppsnet.github.io/
A Dataset of Anatomical Environments for Medical Robots: Modeling Respiratory Deformation
Oct 06, 2023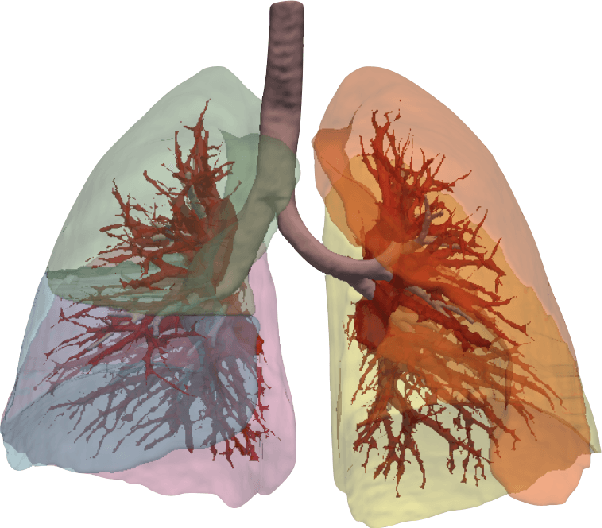
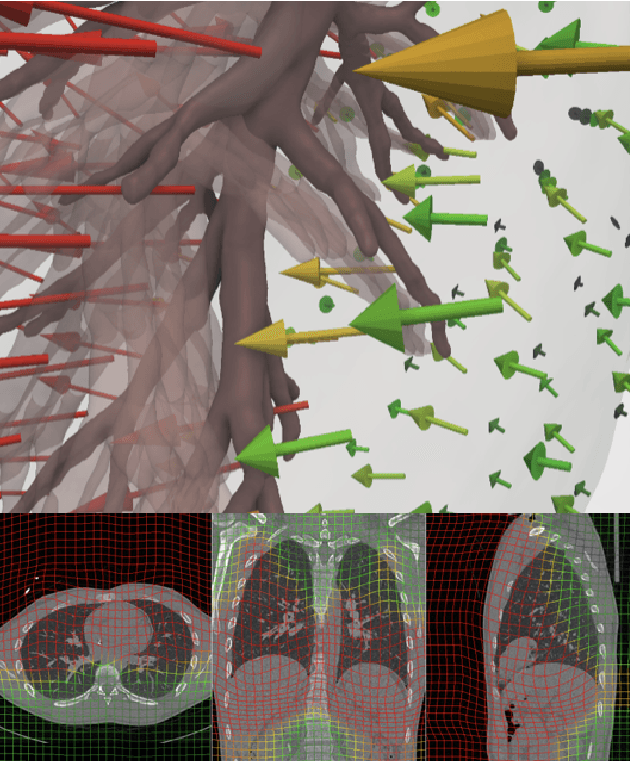
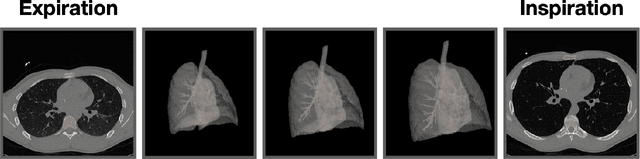
Abstract:Anatomical models of a medical robot's environment can significantly help guide design and development of a new robotic system. These models can be used for benchmarking motion planning algorithms, evaluating controllers, optimizing mechanical design choices, simulating procedures, and even as resources for data generation. Currently, the time-consuming task of generating these environments is repeatedly performed by individual research groups and rarely shared broadly. This not only leads to redundant efforts, but also makes it challenging to compare systems and algorithms accurately. In this work, we present a collection of clinically-relevant anatomical environments for medical robots operating in the lungs. Since anatomical deformation is a fundamental challenge for medical robots operating in the lungs, we describe a way to model respiratory deformation in these environments using patient-derived data. We share the environments and deformation data publicly by adding them to the Medical Robotics Anatomical Dataset (Med-RAD), our public dataset of anatomical environments for medical robots.
Autonomous Medical Needle Steering In Vivo
Nov 04, 2022Abstract:The use of needles to access sites within organs is fundamental to many interventional medical procedures both for diagnosis and treatment. Safe and accurate navigation of a needle through living tissue to an intra-tissue target is currently often challenging or infeasible due to the presence of anatomical obstacles in the tissue, high levels of uncertainty, and natural tissue motion (e.g., due to breathing). Medical robots capable of automating needle-based procedures in vivo have the potential to overcome these challenges and enable an enhanced level of patient care and safety. In this paper, we show the first medical robot that autonomously navigates a needle inside living tissue around anatomical obstacles to an intra-tissue target. Our system leverages an aiming device and a laser-patterned highly flexible steerable needle, a type of needle capable of maneuvering along curvilinear trajectories to avoid obstacles. The autonomous robot accounts for anatomical obstacles and uncertainty in living tissue/needle interaction with replanning and control and accounts for respiratory motion by defining safe insertion time windows during the breathing cycle. We apply the system to lung biopsy, which is critical in the diagnosis of lung cancer, the leading cause of cancer-related death in the United States. We demonstrate successful performance of our system in multiple in vivo porcine studies and also demonstrate that our approach leveraging autonomous needle steering outperforms a standard manual clinical technique for lung nodule access.
A Clinical Dataset for the Evaluation of Motion Planners in Medical Applications
Oct 19, 2022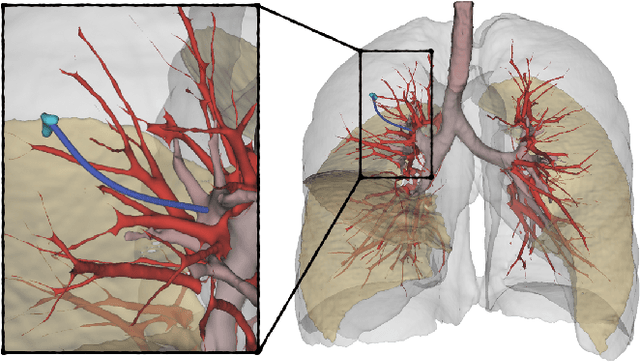
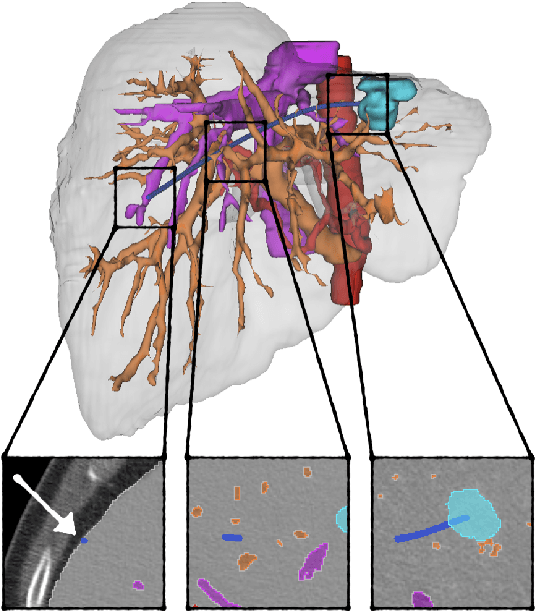
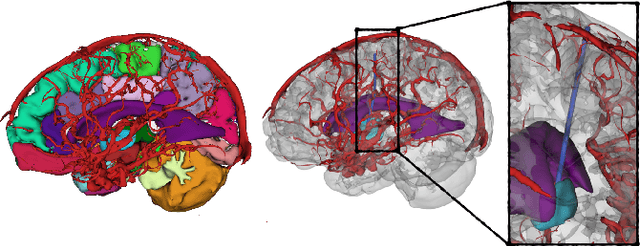
Abstract:The prospect of using autonomous robots to enhance the capabilities of physicians and enable novel procedures has led to considerable efforts in developing medical robots and incorporating autonomous capabilities. Motion planning is a core component for any such system working in an environment that demands near perfect levels of safety, reliability, and precision. Despite the extensive and promising work that has gone into developing motion planners for medical robots, a standardized and clinically-meaningful way to compare existing algorithms and evaluate novel planners and robots is not well established. We present the Medical Motion Planning Dataset (Med-MPD), a publicly-available dataset of real clinical scenarios in various organs for the purpose of evaluating motion planners for minimally-invasive medical robots. Our goal is that this dataset serve as a first step towards creating a larger robust medical motion planning benchmark framework, advance research into medical motion planners, and lift some of the burden of generating medical evaluation data.
Clinical Concept Embeddings Learned from Massive Sources of Multimodal Medical Data
May 18, 2018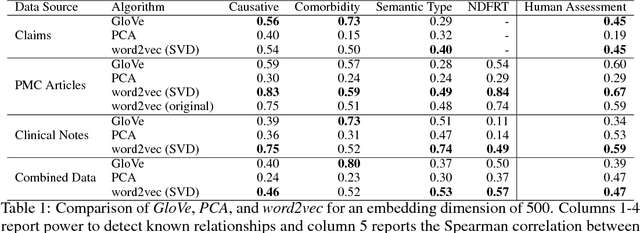
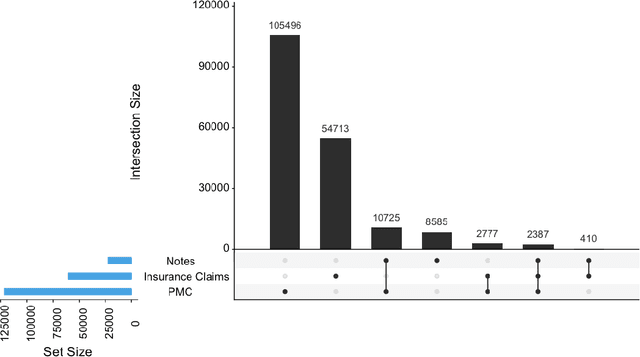
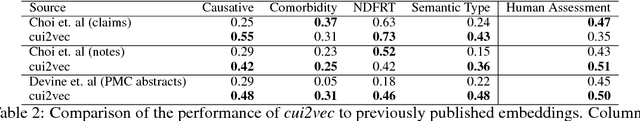
Abstract:Word embeddings are a popular approach to unsupervised learning of word relationships that are widely used in natural language processing. In this article, we present a new set of embeddings for medical concepts learned using an extremely large collection of multimodal medical data. Leaning on recent theoretical insights, we demonstrate how an insurance claims database of 60 million members, a collection of 20 million clinical notes, and 1.7 million full text biomedical journal articles can be combined to embed concepts into a common space, resulting in the largest ever set of embeddings for 108,477 medical concepts. To evaluate our approach, we present a new benchmark methodology based on statistical power specifically designed to test embeddings of medical concepts. Our approach, called cui2vec, attains state of the art performance relative to previous methods in most instances. Finally, we provide a downloadable set of pre-trained embeddings for other researchers to use, as well as an online tool for interactive exploration of the cui2vec embeddings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge