Hector Yee
General Geospatial Inference with a Population Dynamics Foundation Model
Nov 13, 2024



Abstract:Supporting the health and well-being of dynamic populations around the world requires governmental agencies, organizations and researchers to understand and reason over complex relationships between human behavior and local contexts in order to identify high-risk groups and strategically allocate limited resources. Traditional approaches to these classes of problems often entail developing manually curated, task-specific features and models to represent human behavior and the natural and built environment, which can be challenging to adapt to new, or even, related tasks. To address this, we introduce a Population Dynamics Foundation Model (PDFM) that aims to capture the relationships between diverse data modalities and is applicable to a broad range of geospatial tasks. We first construct a geo-indexed dataset for postal codes and counties across the United States, capturing rich aggregated information on human behavior from maps, busyness, and aggregated search trends, and environmental factors such as weather and air quality. We then model this data and the complex relationships between locations using a graph neural network, producing embeddings that can be adapted to a wide range of downstream tasks using relatively simple models. We evaluate the effectiveness of our approach by benchmarking it on 27 downstream tasks spanning three distinct domains: health indicators, socioeconomic factors, and environmental measurements. The approach achieves state-of-the-art performance on all 27 geospatial interpolation tasks, and on 25 out of the 27 extrapolation and super-resolution tasks. We combined the PDFM with a state-of-the-art forecasting foundation model, TimesFM, to predict unemployment and poverty, achieving performance that surpasses fully supervised forecasting. The full set of embeddings and sample code are publicly available for researchers.
Community search signatures as foundation features for human-centered geospatial modeling
Oct 30, 2024



Abstract:Aggregated relative search frequencies offer a unique composite signal reflecting people's habits, concerns, interests, intents, and general information needs, which are not found in other readily available datasets. Temporal search trends have been successfully used in time series modeling across a variety of domains such as infectious diseases, unemployment rates, and retail sales. However, most existing applications require curating specialized datasets of individual keywords, queries, or query clusters, and the search data need to be temporally aligned with the outcome variable of interest. We propose a novel approach for generating an aggregated and anonymized representation of search interest as foundation features at the community level for geospatial modeling. We benchmark these features using spatial datasets across multiple domains. In zip codes with a population greater than 3000 that cover over 95% of the contiguous US population, our models for predicting missing values in a 20% set of holdout counties achieve an average $R^2$ score of 0.74 across 21 health variables, and 0.80 across 6 demographic and environmental variables. Our results demonstrate that these search features can be used for spatial predictions without strict temporal alignment, and that the resulting models outperform spatial interpolation and state of the art methods using satellite imagery features.
Scalable and accurate deep learning for electronic health records
May 11, 2018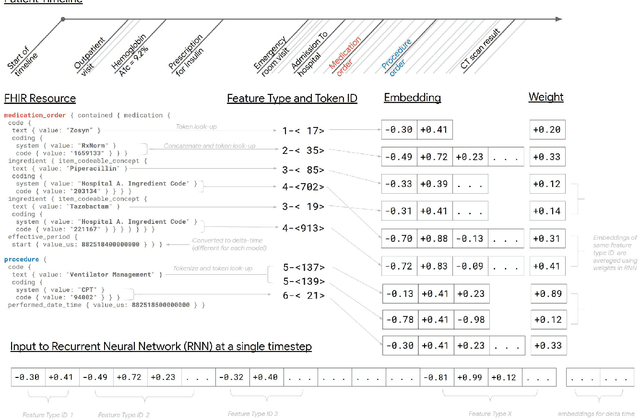
Abstract:Predictive modeling with electronic health record (EHR) data is anticipated to drive personalized medicine and improve healthcare quality. Constructing predictive statistical models typically requires extraction of curated predictor variables from normalized EHR data, a labor-intensive process that discards the vast majority of information in each patient's record. We propose a representation of patients' entire, raw EHR records based on the Fast Healthcare Interoperability Resources (FHIR) format. We demonstrate that deep learning methods using this representation are capable of accurately predicting multiple medical events from multiple centers without site-specific data harmonization. We validated our approach using de-identified EHR data from two U.S. academic medical centers with 216,221 adult patients hospitalized for at least 24 hours. In the sequential format we propose, this volume of EHR data unrolled into a total of 46,864,534,945 data points, including clinical notes. Deep learning models achieved high accuracy for tasks such as predicting in-hospital mortality (AUROC across sites 0.93-0.94), 30-day unplanned readmission (AUROC 0.75-0.76), prolonged length of stay (AUROC 0.85-0.86), and all of a patient's final discharge diagnoses (frequency-weighted AUROC 0.90). These models outperformed state-of-the-art traditional predictive models in all cases. We also present a case-study of a neural-network attribution system, which illustrates how clinicians can gain some transparency into the predictions. We believe that this approach can be used to create accurate and scalable predictions for a variety of clinical scenarios, complete with explanations that directly highlight evidence in the patient's chart.
* Published version from https://www.nature.com/articles/s41746-018-0029-1
Affinity Weighted Embedding
Jan 17, 2013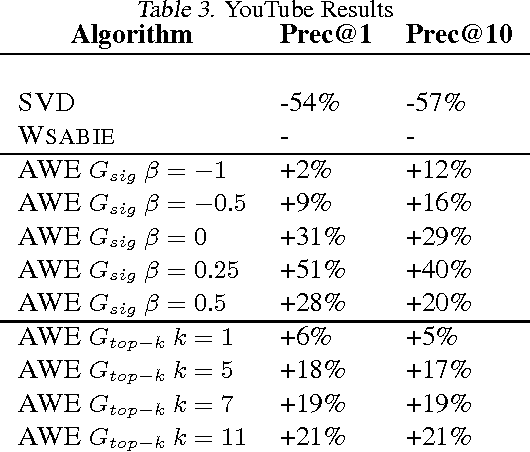
Abstract:Supervised (linear) embedding models like Wsabie and PSI have proven successful at ranking, recommendation and annotation tasks. However, despite being scalable to large datasets they do not take full advantage of the extra data due to their linear nature, and typically underfit. We propose a new class of models which aim to provide improved performance while retaining many of the benefits of the existing class of embedding models. Our new approach works by iteratively learning a linear embedding model where the next iteration's features and labels are reweighted as a function of the previous iteration. We describe several variants of the family, and give some initial results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge