Peggy Bui
LLM-based Text Simplification and its Effect on User Comprehension and Cognitive Load
May 04, 2025Abstract:Information on the web, such as scientific publications and Wikipedia, often surpasses users' reading level. To help address this, we used a self-refinement approach to develop a LLM capability for minimally lossy text simplification. To validate our approach, we conducted a randomized study involving 4563 participants and 31 texts spanning 6 broad subject areas: PubMed (biomedical scientific articles), biology, law, finance, literature/philosophy, and aerospace/computer science. Participants were randomized to viewing original or simplified texts in a subject area, and answered multiple-choice questions (MCQs) that tested their comprehension of the text. The participants were also asked to provide qualitative feedback such as task difficulty. Our results indicate that participants who read the simplified text answered more MCQs correctly than their counterparts who read the original text (3.9% absolute increase, p<0.05). This gain was most striking with PubMed (14.6%), while more moderate gains were observed for finance (5.5%), aerospace/computer science (3.8%) domains, and legal (3.5%). Notably, the results were robust to whether participants could refer back to the text while answering MCQs. The absolute accuracy decreased by up to ~9% for both original and simplified setups where participants could not refer back to the text, but the ~4% overall improvement persisted. Finally, participants' self-reported perceived ease based on a simplified NASA Task Load Index was greater for those who read the simplified text (absolute change on a 5-point scale 0.33, p<0.05). This randomized study, involving an order of magnitude more participants than prior works, demonstrates the potential of LLMs to make complex information easier to understand. Our work aims to enable a broader audience to better learn and make use of expert knowledge available on the web, improving information accessibility.
Closing the AI generalization gap by adjusting for dermatology condition distribution differences across clinical settings
Feb 23, 2024Abstract:Recently, there has been great progress in the ability of artificial intelligence (AI) algorithms to classify dermatological conditions from clinical photographs. However, little is known about the robustness of these algorithms in real-world settings where several factors can lead to a loss of generalizability. Understanding and overcoming these limitations will permit the development of generalizable AI that can aid in the diagnosis of skin conditions across a variety of clinical settings. In this retrospective study, we demonstrate that differences in skin condition distribution, rather than in demographics or image capture mode are the main source of errors when an AI algorithm is evaluated on data from a previously unseen source. We demonstrate a series of steps to close this generalization gap, requiring progressively more information about the new source, ranging from the condition distribution to training data enriched for data less frequently seen during training. Our results also suggest comparable performance from end-to-end fine tuning versus fine tuning solely the classification layer on top of a frozen embedding model. Our approach can inform the adaptation of AI algorithms to new settings, based on the information and resources available.
Does Your Dermatology Classifier Know What It Doesn't Know? Detecting the Long-Tail of Unseen Conditions
Apr 08, 2021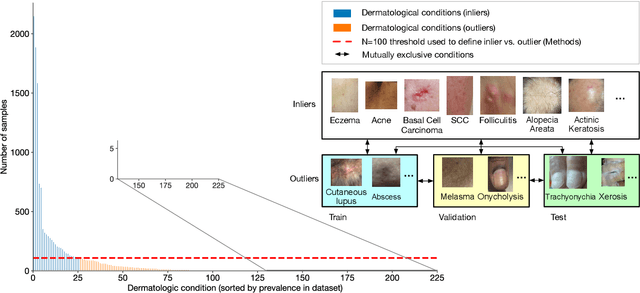
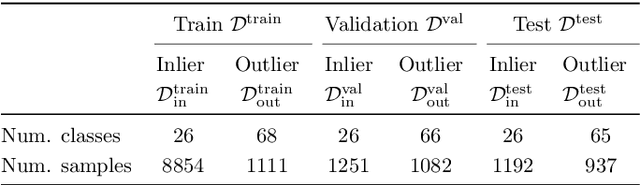
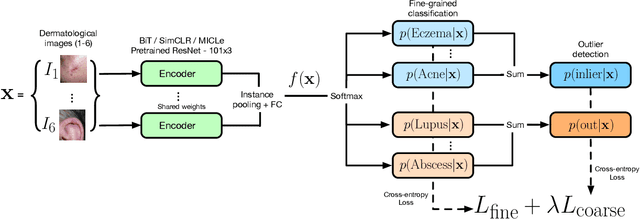
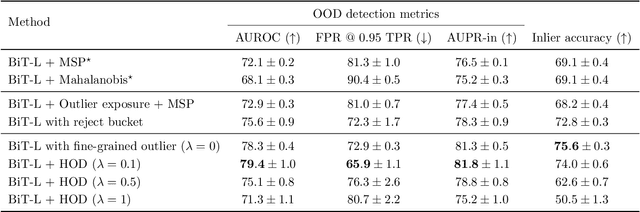
Abstract:We develop and rigorously evaluate a deep learning based system that can accurately classify skin conditions while detecting rare conditions for which there is not enough data available for training a confident classifier. We frame this task as an out-of-distribution (OOD) detection problem. Our novel approach, hierarchical outlier detection (HOD) assigns multiple abstention classes for each training outlier class and jointly performs a coarse classification of inliers vs. outliers, along with fine-grained classification of the individual classes. We demonstrate the effectiveness of the HOD loss in conjunction with modern representation learning approaches (BiT, SimCLR, MICLe) and explore different ensembling strategies for further improving the results. We perform an extensive subgroup analysis over conditions of varying risk levels and different skin types to investigate how the OOD detection performance changes over each subgroup and demonstrate the gains of our framework in comparison to baselines. Finally, we introduce a cost metric to approximate downstream clinical impact. We use this cost metric to compare the proposed method against a baseline system, thereby making a stronger case for the overall system effectiveness in a real-world deployment scenario.
Supervised Transfer Learning at Scale for Medical Imaging
Jan 21, 2021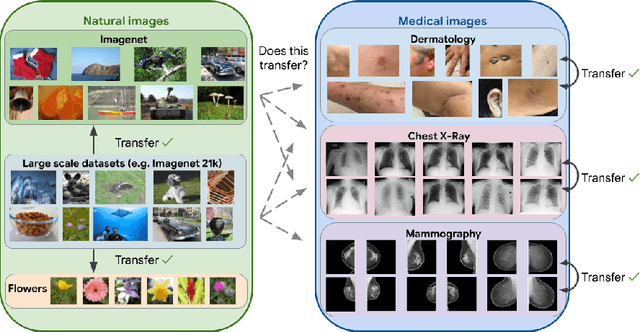
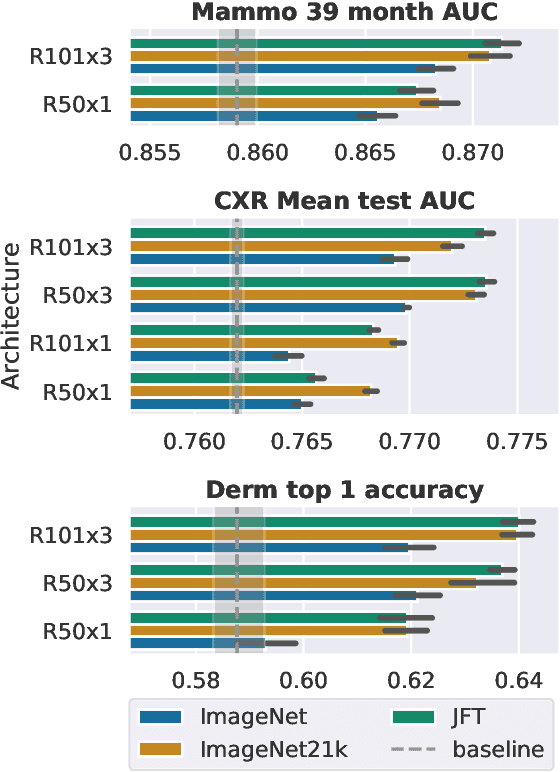
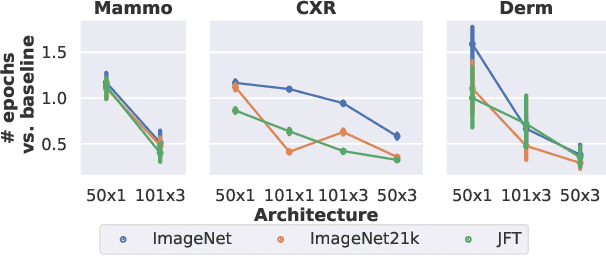
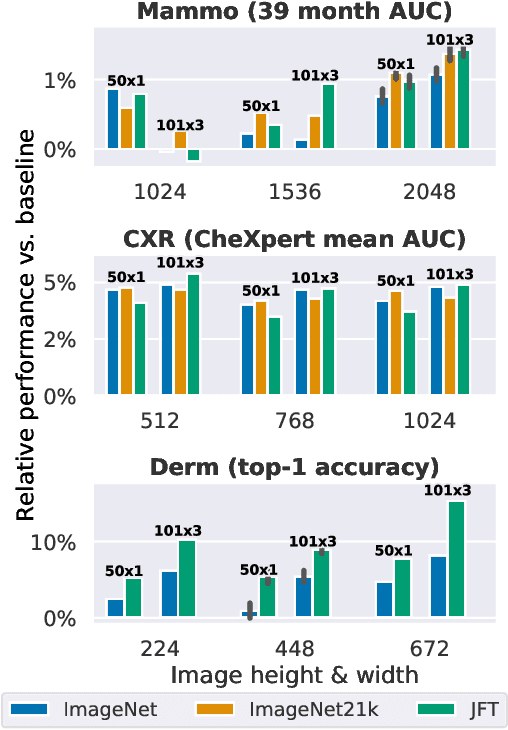
Abstract:Transfer learning is a standard technique to improve performance on tasks with limited data. However, for medical imaging, the value of transfer learning is less clear. This is likely due to the large domain mismatch between the usual natural-image pre-training (e.g. ImageNet) and medical images. However, recent advances in transfer learning have shown substantial improvements from scale. We investigate whether modern methods can change the fortune of transfer learning for medical imaging. For this, we study the class of large-scale pre-trained networks presented by Kolesnikov et al. on three diverse imaging tasks: chest radiography, mammography, and dermatology. We study both transfer performance and critical properties for the deployment in the medical domain, including: out-of-distribution generalization, data-efficiency, sub-group fairness, and uncertainty estimation. Interestingly, we find that for some of these properties transfer from natural to medical images is indeed extremely effective, but only when performed at sufficient scale.
A deep learning system for differential diagnosis of skin diseases
Sep 11, 2019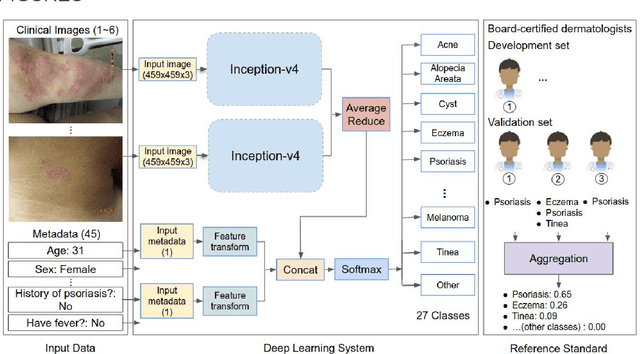
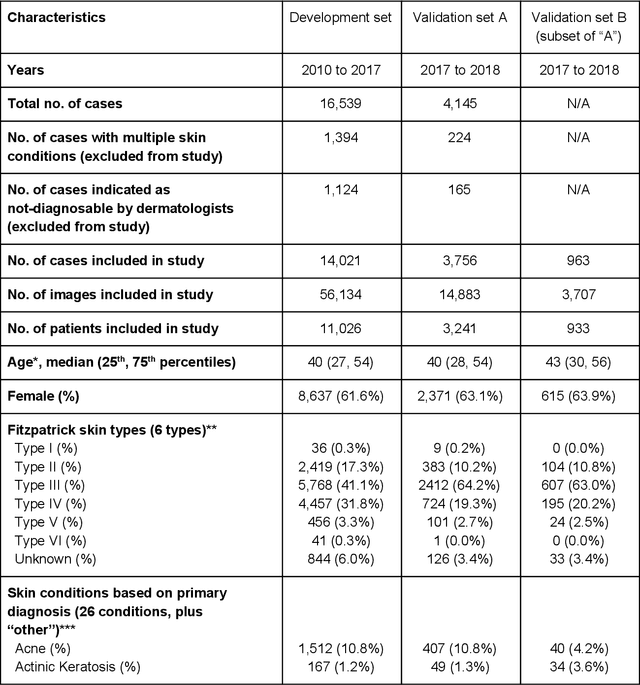
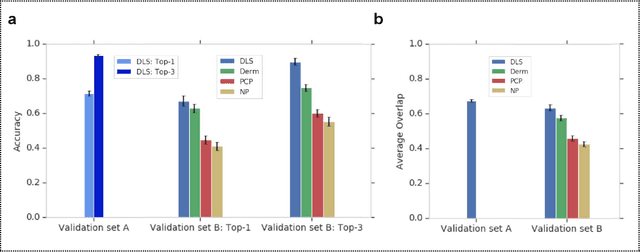
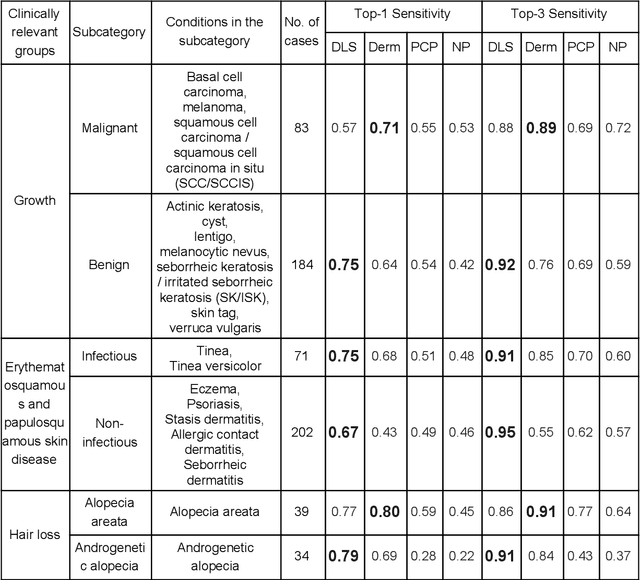
Abstract:Skin conditions affect an estimated 1.9 billion people worldwide. A shortage of dermatologists causes long wait times and leads patients to seek dermatologic care from general practitioners. However, the diagnostic accuracy of general practitioners has been reported to be only 0.24-0.70 (compared to 0.77-0.96 for dermatologists), resulting in referral errors, delays in care, and errors in diagnosis and treatment. In this paper, we developed a deep learning system (DLS) to provide a differential diagnosis of skin conditions for clinical cases (skin photographs and associated medical histories). The DLS distinguishes between 26 skin conditions that represent roughly 80% of the volume of skin conditions seen in primary care. The DLS was developed and validated using de-identified cases from a teledermatology practice serving 17 clinical sites via a temporal split: the first 14,021 cases for development and the last 3,756 cases for validation. On the validation set, where a panel of three board-certified dermatologists defined the reference standard for every case, the DLS achieved 0.71 and 0.93 top-1 and top-3 accuracies respectively. For a random subset of the validation set (n=963 cases), 18 clinicians reviewed the cases for comparison. On this subset, the DLS achieved a 0.67 top-1 accuracy, non-inferior to board-certified dermatologists (0.63, p<0.001), and higher than primary care physicians (PCPs, 0.45) and nurse practitioners (NPs, 0.41). The top-3 accuracy showed a similar trend: 0.90 DLS, 0.75 dermatologists, 0.60 PCPs, and 0.55 NPs. These results highlight the potential of the DLS to augment general practitioners to accurately diagnose skin conditions by suggesting differential diagnoses that may not have been considered. Future work will be needed to prospectively assess the clinical impact of using this tool in actual clinical workflows.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge