Atilla P. Kiraly
Health AI Developer Foundations
Nov 26, 2024



Abstract:Robust medical Machine Learning (ML) models have the potential to revolutionize healthcare by accelerating clinical research, improving workflows and outcomes, and producing novel insights or capabilities. Developing such ML models from scratch is cost prohibitive and requires substantial compute, data, and time (e.g., expert labeling). To address these challenges, we introduce Health AI Developer Foundations (HAI-DEF), a suite of pre-trained, domain-specific foundation models, tools, and recipes to accelerate building ML for health applications. The models cover various modalities and domains, including radiology (X-rays and computed tomography), histopathology, dermatological imaging, and audio. These models provide domain specific embeddings that facilitate AI development with less labeled data, shorter training times, and reduced computational costs compared to traditional approaches. In addition, we utilize a common interface and style across these models, and prioritize usability to enable developers to integrate HAI-DEF efficiently. We present model evaluations across various tasks and conclude with a discussion of their application and evaluation, covering the importance of ensuring efficacy, fairness, and equity. Finally, while HAI-DEF and specifically the foundation models lower the barrier to entry for ML in healthcare, we emphasize the importance of validation with problem- and population-specific data for each desired usage setting. This technical report will be updated over time as more modalities and features are added.
Deep Learning for Distinguishing Normal versus Abnormal Chest Radiographs and Generalization to Unseen Diseases
Oct 22, 2020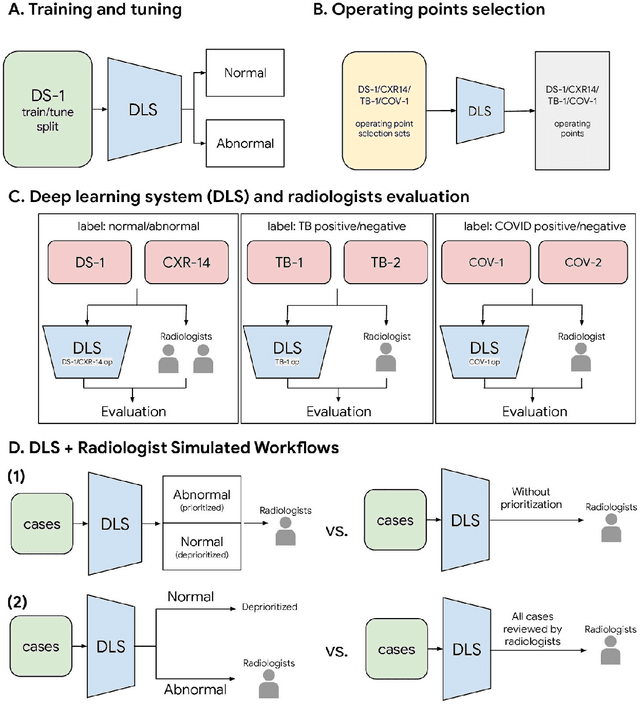
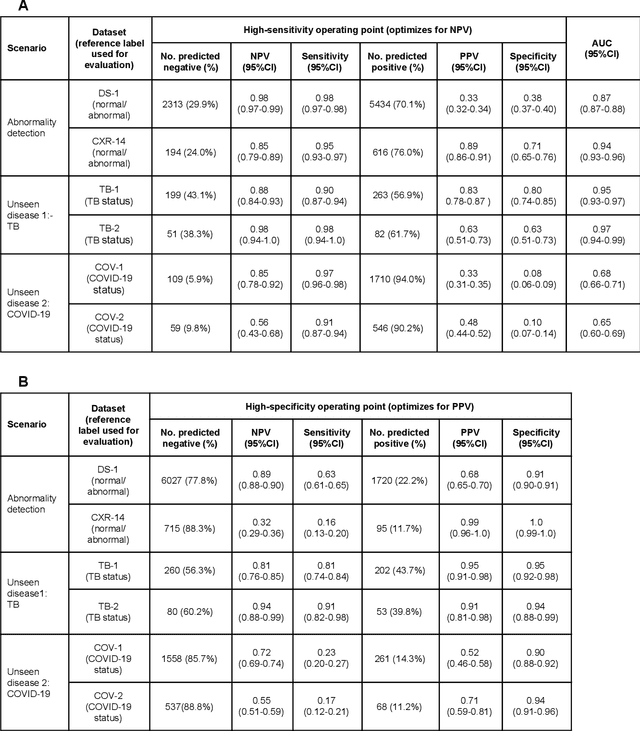
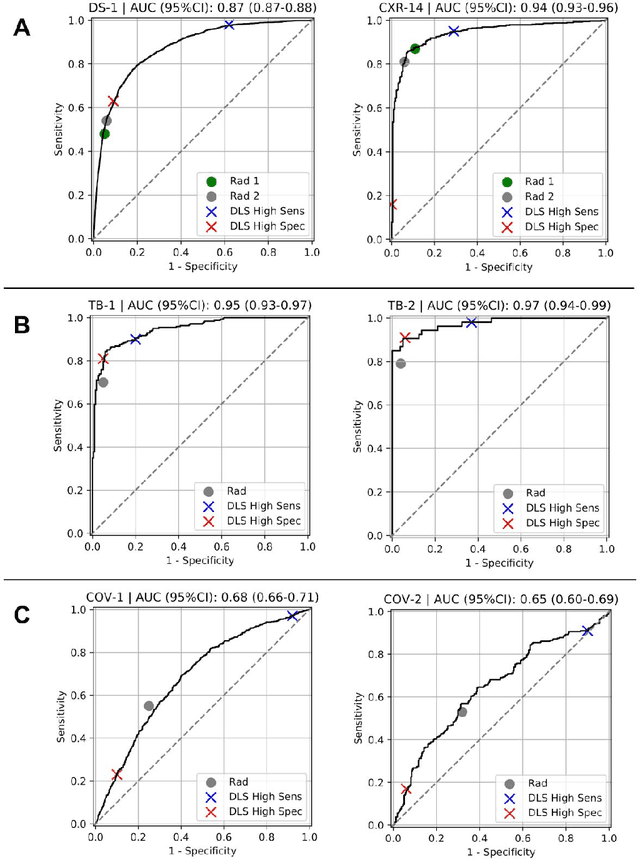
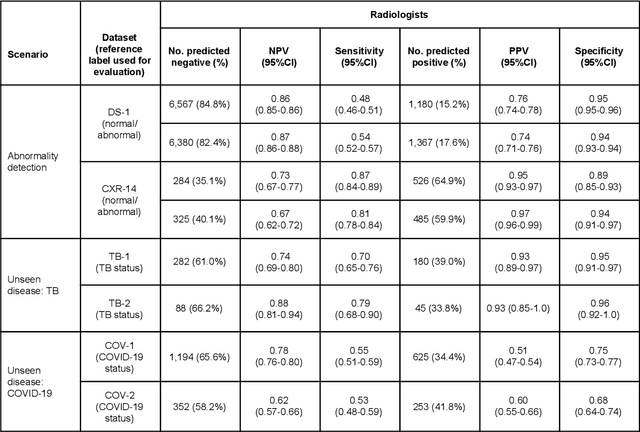
Abstract:Chest radiography (CXR) is the most widely-used thoracic clinical imaging modality and is crucial for guiding the management of cardiothoracic conditions. The detection of specific CXR findings has been the main focus of several artificial intelligence (AI) systems. However, the wide range of possible CXR abnormalities makes it impractical to build specific systems to detect every possible condition. In this work, we developed and evaluated an AI system to classify CXRs as normal or abnormal. For development, we used a de-identified dataset of 248,445 patients from a multi-city hospital network in India. To assess generalizability, we evaluated our system using 6 international datasets from India, China, and the United States. Of these datasets, 4 focused on diseases that the AI was not trained to detect: 2 datasets with tuberculosis and 2 datasets with coronavirus disease 2019. Our results suggest that the AI system generalizes to new patient populations and abnormalities. In a simulated workflow where the AI system prioritized abnormal cases, the turnaround time for abnormal cases reduced by 7-28%. These results represent an important step towards evaluating whether AI can be safely used to flag cases in a general setting where previously unseen abnormalities exist.
Deep Adversarial Context-Aware Landmark Detection for Ultrasound Imaging
May 28, 2018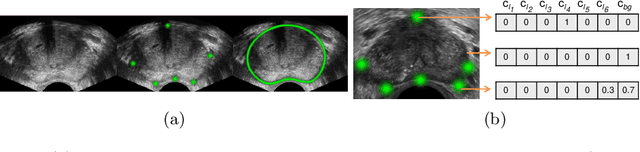

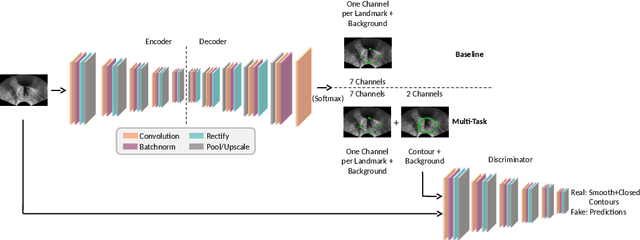
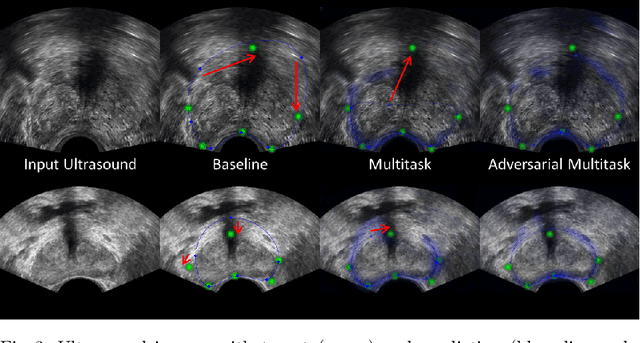
Abstract:Real-time localization of prostate gland in trans-rectal ultrasound images is a key technology that is required to automate the ultrasound guided prostate biopsy procedures. In this paper, we propose a new deep learning based approach which is aimed at localizing several prostate landmarks efficiently and robustly. We propose a multitask learning approach primarily to make the overall algorithm more contextually aware. In this approach, we not only consider the explicit learning of landmark locations, but also build-in a mechanism to learn the contour of the prostate. This multitask learning is further coupled with an adversarial arm to promote the generation of feasible structures. We have trained this network using ~4000 labeled trans-rectal ultrasound images and tested on an independent set of images with ground truth landmark locations. We have achieved an overall Dice score of 92.6% for the adversarially trained multitask approach, which is significantly better than the Dice score of 88.3% obtained by only learning of landmark locations. The overall mean distance error using the adversarial multitask approach has also improved by 20% while reducing the standard deviation of the error compared to learning landmark locations only. In terms of computational complexity both approaches can process the images in real-time using standard computer with a standard CUDA enabled GPU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge