Rethinking Transformer for Long Contextual Histopathology Whole Slide Image Analysis
Paper and Code
Oct 18, 2024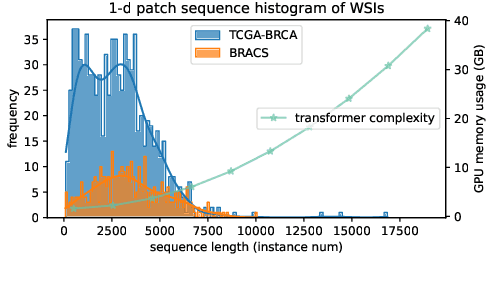
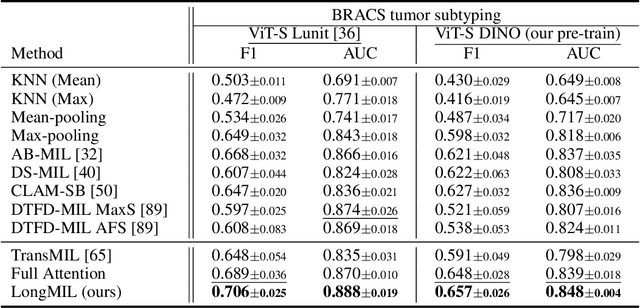
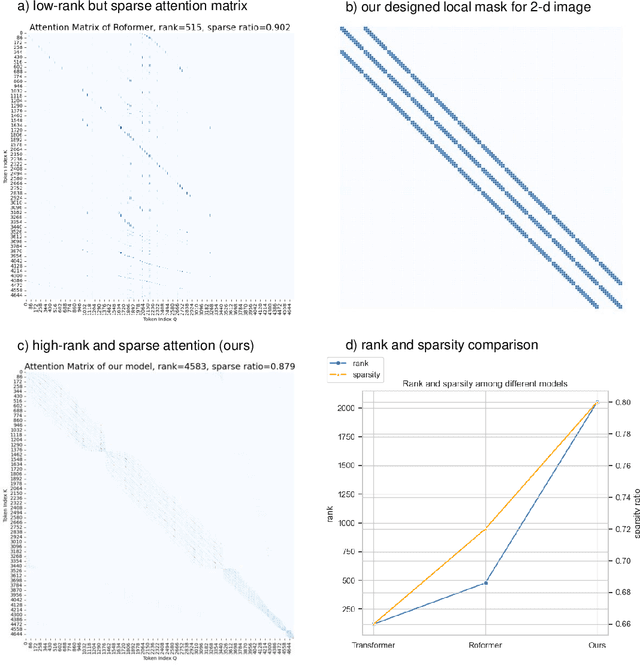
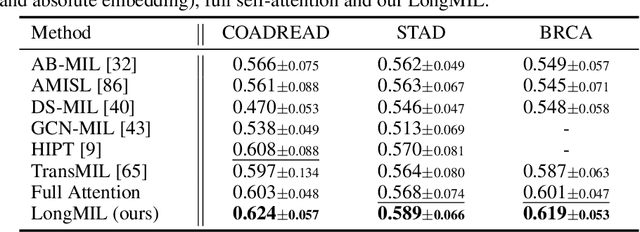
Histopathology Whole Slide Image (WSI) analysis serves as the gold standard for clinical cancer diagnosis in the daily routines of doctors. To develop computer-aided diagnosis model for WSIs, previous methods typically employ Multi-Instance Learning to enable slide-level prediction given only slide-level labels. Among these models, vanilla attention mechanisms without pairwise interactions have traditionally been employed but are unable to model contextual information. More recently, self-attention models have been utilized to address this issue. To alleviate the computational complexity of long sequences in large WSIs, methods like HIPT use region-slicing, and TransMIL employs approximation of full self-attention. Both approaches suffer from suboptimal performance due to the loss of key information. Moreover, their use of absolute positional embedding struggles to effectively handle long contextual dependencies in shape-varying WSIs. In this paper, we first analyze how the low-rank nature of the long-sequence attention matrix constrains the representation ability of WSI modelling. Then, we demonstrate that the rank of attention matrix can be improved by focusing on local interactions via a local attention mask. Our analysis shows that the local mask aligns with the attention patterns in the lower layers of the Transformer. Furthermore, the local attention mask can be implemented during chunked attention calculation, reducing the quadratic computational complexity to linear with a small local bandwidth. Building on this, we propose a local-global hybrid Transformer for both computational acceleration and local-global information interactions modelling. Our method, Long-contextual MIL (LongMIL), is evaluated through extensive experiments on various WSI tasks to validate its superiority. Our code will be available at github.com/invoker-LL/Long-MIL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge