Minhao Hu
Cycle-Consistent Bridge Diffusion Model for Accelerated MRI Reconstruction
Dec 13, 2024



Abstract:Accelerated MRI reconstruction techniques aim to reduce examination time while maintaining high image fidelity, which is highly desirable in clinical settings for improving patient comfort and hospital efficiency. Existing deep learning methods typically reconstruct images from under-sampled data with traditional reconstruction approaches, but they still struggle to provide high-fidelity results. Diffusion models show great potential to improve fidelity of generated images in recent years. However, their inference process starting with a random Gaussian noise introduces instability into the results and usually requires thousands of sampling steps, resulting in sub-optimal reconstruction quality and low efficiency. To address these challenges, we propose Cycle-Consistent Bridge Diffusion Model (CBDM). CBDM employs two bridge diffusion models to construct a cycle-consistent diffusion process with a consistency loss, enhancing the fine-grained details of reconstructed images and reducing the number of diffusion steps. Moreover, CBDM incorporates a Contourlet Decomposition Embedding Module (CDEM) which captures multi-scale structural texture knowledge in images through frequency domain decomposition pyramids and directional filter banks to improve structural fidelity. Extensive experiments demonstrate the superiority of our model by higher reconstruction quality and fewer training iterations, achieving a new state of the art for accelerated MRI reconstruction in both fastMRI and IXI datasets.
Cross Group Attention and Group-wise Rolling for Multimodal Medical Image Synthesis
Nov 22, 2024Abstract:Multimodal MR image synthesis aims to generate missing modality image by fusing and mapping a few available MRI data. Most existing approaches typically adopt an image-to-image translation scheme. However, these methods often suffer from sub-optimal performance due to the spatial misalignment between different modalities while they are typically treated as input channels. Therefore, in this paper, we propose an Adaptive Group-wise Interaction Network (AGI-Net) that explores both inter-modality and intra-modality relationships for multimodal MR image synthesis. Specifically, groups are first pre-defined along the channel dimension and then we perform an adaptive rolling for the standard convolutional kernel to capture inter-modality spatial correspondences. At the same time, a cross-group attention module is introduced to fuse information across different channel groups, leading to better feature representation. We evaluated the effectiveness of our model on the publicly available IXI and BraTS2023 datasets, where the AGI-Net achieved state-of-the-art performance for multimodal MR image synthesis. Code will be released.
Scribble-Supervised Medical Image Segmentation via Dual-Branch Network and Dynamically Mixed Pseudo Labels Supervision
Mar 04, 2022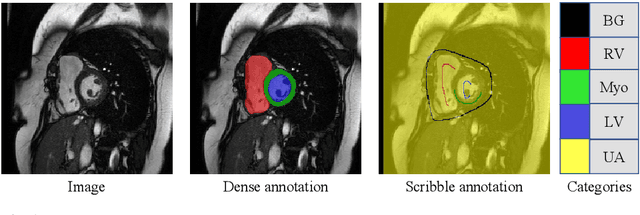
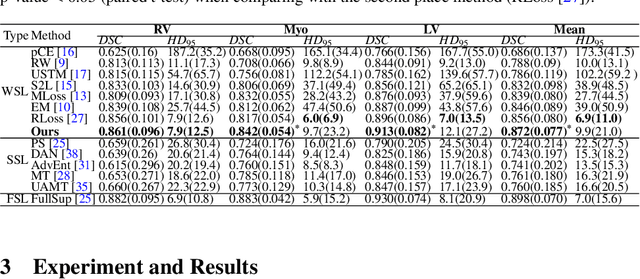
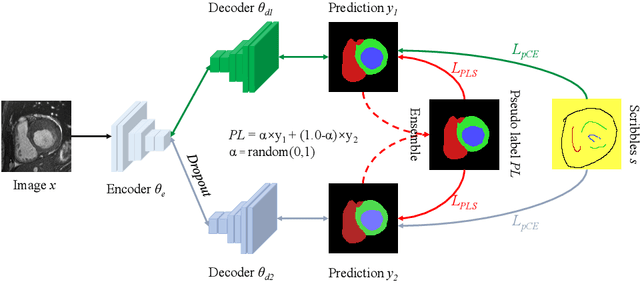
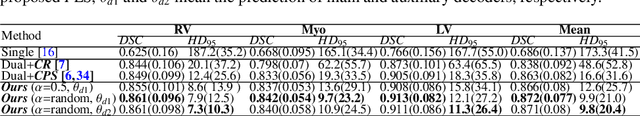
Abstract:Medical image segmentation plays an irreplaceable role in computer-assisted diagnosis, treatment planning, and following-up. Collecting and annotating a large-scale dataset is crucial to training a powerful segmentation model, but producing high-quality segmentation masks is an expensive and time-consuming procedure. Recently, weakly-supervised learning that uses sparse annotations (points, scribbles, bounding boxes) for network training has achieved encouraging performance and shown the potential for annotation cost reduction. However, due to the limited supervision signal of sparse annotations, it is still challenging to employ them for networks training directly. In this work, we propose a simple yet efficient scribble-supervised image segmentation method and apply it to cardiac MRI segmentation. Specifically, we employ a dual-branch network with one encoder and two slightly different decoders for image segmentation and dynamically mix the two decoders' predictions to generate pseudo labels for auxiliary supervision. By combining the scribble supervision and auxiliary pseudo labels supervision, the dual-branch network can efficiently learn from scribble annotations end-to-end. Experiments on the public ACDC dataset show that our method performs better than current scribble-supervised segmentation methods and also outperforms several semi-supervised segmentation methods.
Semi-Supervised Medical Image Segmentation via Cross Teaching between CNN and Transformer
Dec 09, 2021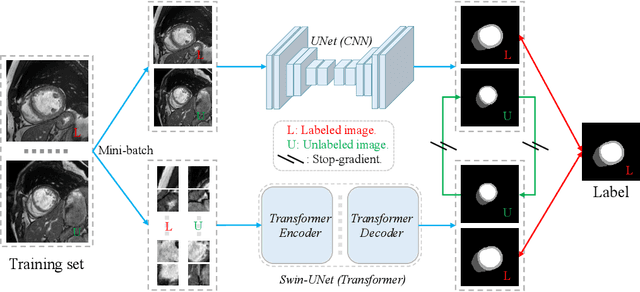
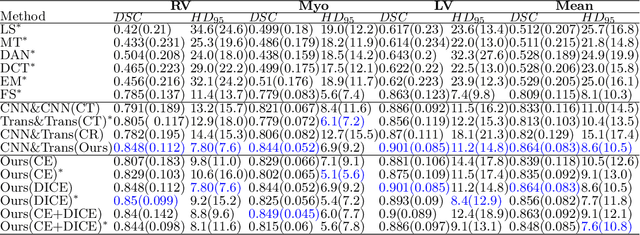

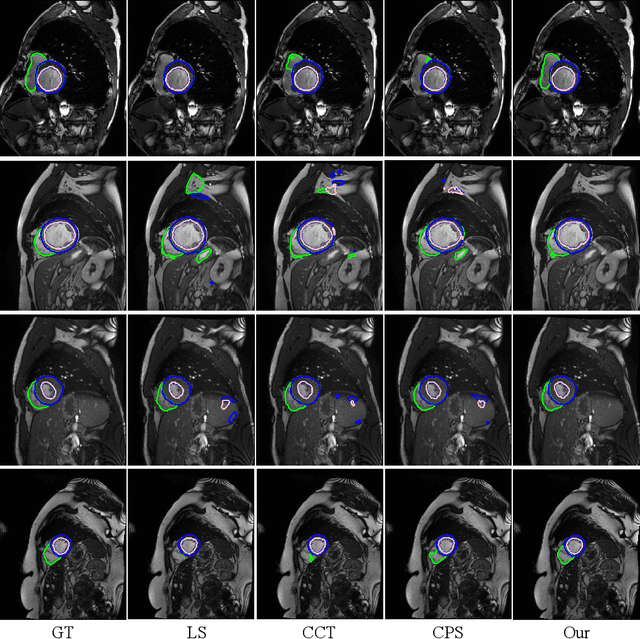
Abstract:Recently, deep learning with Convolutional Neural Networks (CNNs) and Transformers has shown encouraging results in fully supervised medical image segmentation. However, it is still challenging for them to achieve good performance with limited annotations for training. In this work, we present a very simple yet efficient framework for semi-supervised medical image segmentation by introducing the cross teaching between CNN and Transformer. Specifically, we simplify the classical deep co-training from consistency regularization to cross teaching, where the prediction of a network is used as the pseudo label to supervise the other network directly end-to-end. Considering the difference in learning paradigm between CNN and Transformer, we introduce the Cross Teaching between CNN and Transformer rather than just using CNNs. Experiments on a public benchmark show that our method outperforms eight existing semi-supervised learning methods just with a simpler framework. Notably, this work may be the first attempt to combine CNN and transformer for semi-supervised medical image segmentation and achieve promising results on a public benchmark. The code will be released at: https://github.com/HiLab-git/SSL4MIS.
Knowledge distillation from multi-modal to mono-modal segmentation networks
Jun 17, 2021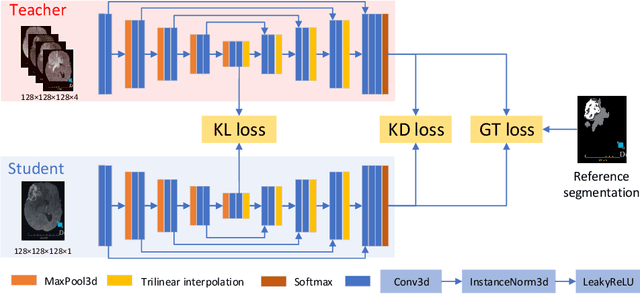
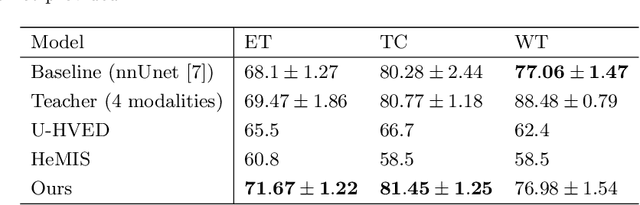
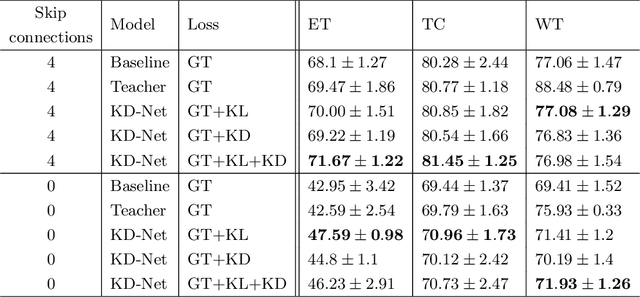
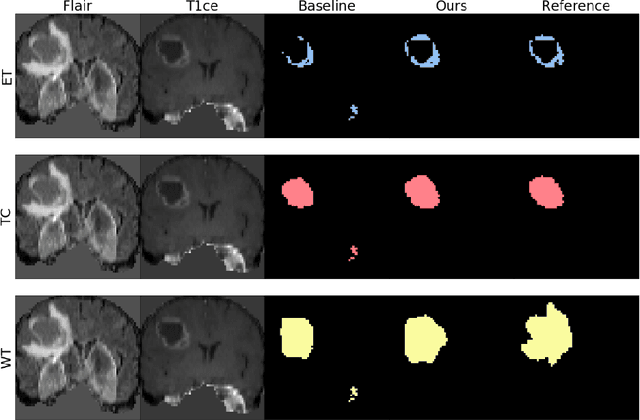
Abstract:The joint use of multiple imaging modalities for medical image segmentation has been widely studied in recent years. The fusion of information from different modalities has demonstrated to improve the segmentation accuracy, with respect to mono-modal segmentations, in several applications. However, acquiring multiple modalities is usually not possible in a clinical setting due to a limited number of physicians and scanners, and to limit costs and scan time. Most of the time, only one modality is acquired. In this paper, we propose KD-Net, a framework to transfer knowledge from a trained multi-modal network (teacher) to a mono-modal one (student). The proposed method is an adaptation of the generalized distillation framework where the student network is trained on a subset (1 modality) of the teacher's inputs (n modalities). We illustrate the effectiveness of the proposed framework in brain tumor segmentation with the BraTS 2018 dataset. Using different architectures, we show that the student network effectively learns from the teacher and always outperforms the baseline mono-modal network in terms of segmentation accuracy.
* MICCAI 2020
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge