Giammarco La Barbera
First Order Logic with Fuzzy Semantics for Describing and Recognizing Nerves in Medical Images
Apr 30, 2025Abstract:This article deals with the description and recognition of fiber bundles, in particular nerves, in medical images, based on the anatomical description of the fiber trajectories. To this end, we propose a logical formalization of this anatomical knowledge. The intrinsically imprecise description of nerves, as found in anatomical textbooks, leads us to propose fuzzy semantics combined with first-order logic. We define a language representing spatial entities, relations between these entities and quantifiers. A formula in this language is then a formalization of the natural language description. The semantics are given by fuzzy representations in a concrete domain and satisfaction degrees of relations. Based on this formalization, a spatial reasoning algorithm is proposed for segmentation and recognition of nerves from anatomical and diffusion magnetic resonance images, which is illustrated on pelvic nerves in pediatric imaging, enabling surgeons to plan surgery.
Anatomically constrained CT image translation for heterogeneous blood vessel segmentation
Oct 04, 2022

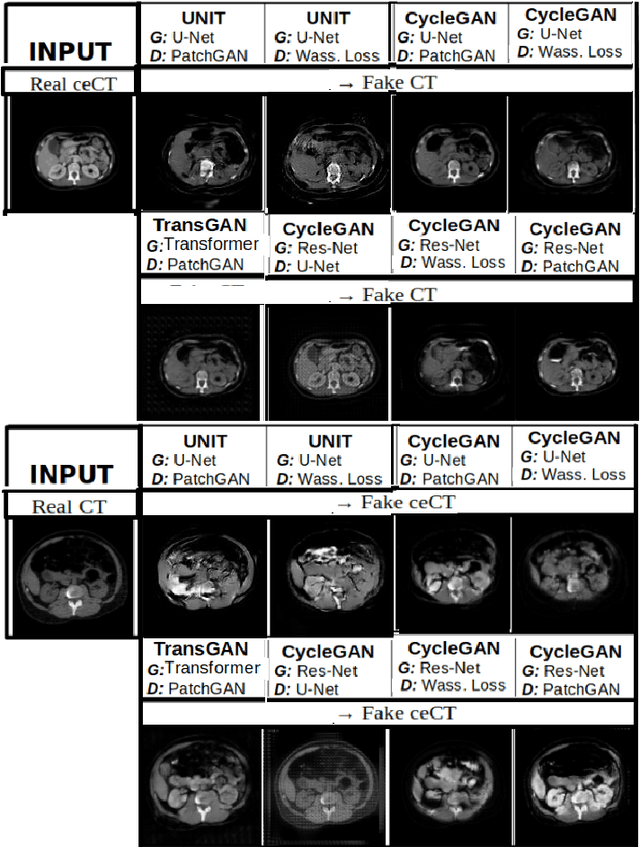
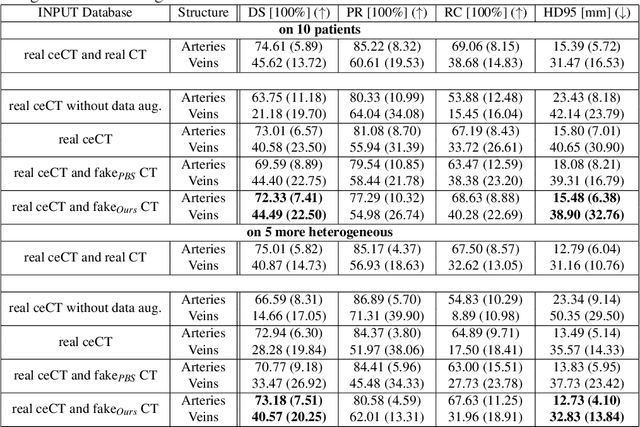
Abstract:Anatomical structures such as blood vessels in contrast-enhanced CT (ceCT) images can be challenging to segment due to the variability in contrast medium diffusion. The combined use of ceCT and contrast-free (CT) CT images can improve the segmentation performances, but at the cost of a double radiation exposure. To limit the radiation dose, generative models could be used to synthesize one modality, instead of acquiring it. The CycleGAN approach has recently attracted particular attention because it alleviates the need for paired data that are difficult to obtain. Despite the great performances demonstrated in the literature, limitations still remain when dealing with 3D volumes generated slice by slice from unpaired datasets with different fields of view. We present an extension of CycleGAN to generate high fidelity images, with good structural consistency, in this context. We leverage anatomical constraints and automatic region of interest selection by adapting the Self-Supervised Body Regressor. These constraints enforce anatomical consistency and allow feeding anatomically-paired input images to the algorithm. Results show qualitative and quantitative improvements, compared to stateof-the-art methods, on the translation task between ceCT and CT images (and vice versa).
Automatic size and pose homogenization with spatial transformer network to improve and accelerate pediatric segmentation
Jul 06, 2021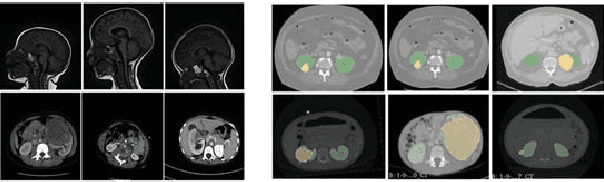

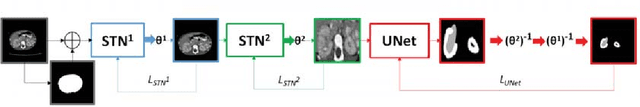
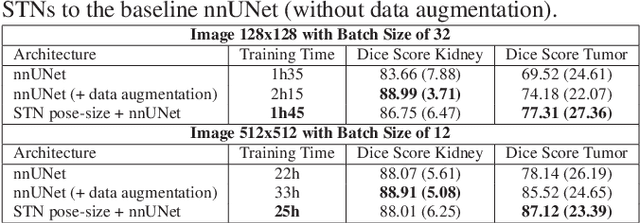
Abstract:Due to a high heterogeneity in pose and size and to a limited number of available data, segmentation of pediatric images is challenging for deep learning methods. In this work, we propose a new CNN architecture that is pose and scale invariant thanks to the use of Spatial Transformer Network (STN). Our architecture is composed of three sequential modules that are estimated together during training: (i) a regression module to estimate a similarity matrix to normalize the input image to a reference one; (ii) a differentiable module to find the region of interest to segment; (iii) a segmentation module, based on the popular UNet architecture, to delineate the object. Unlike the original UNet, which strives to learn a complex mapping, including pose and scale variations, from a finite training dataset, our segmentation module learns a simpler mapping focusing on images with normalized pose and size. Furthermore, the use of an automatic bounding box detection through STN allows saving time and especially memory, while keeping similar performance. We test the proposed method in kidney and renal tumor segmentation on abdominal pediatric CT scanners. Results indicate that the estimated STN homogenization of size and pose accelerates the segmentation (25h), compared to standard data-augmentation (33h), while obtaining a similar quality for the kidney (88.01\% of Dice score) and improving the renal tumor delineation (from 85.52\% to 87.12\%).
* ISBI 2021
Knowledge distillation from multi-modal to mono-modal segmentation networks
Jun 17, 2021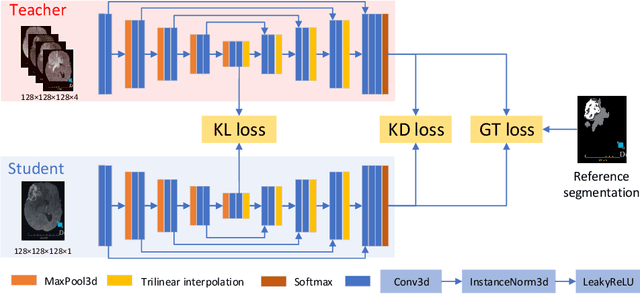
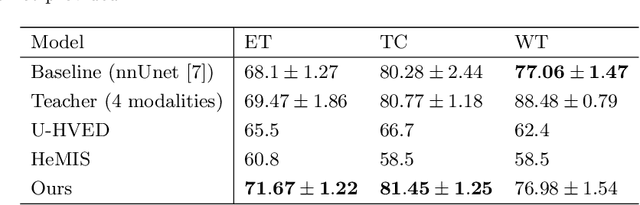
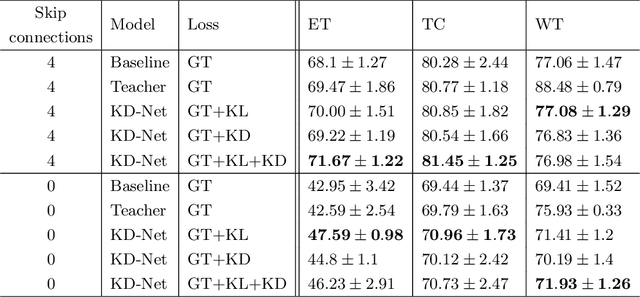
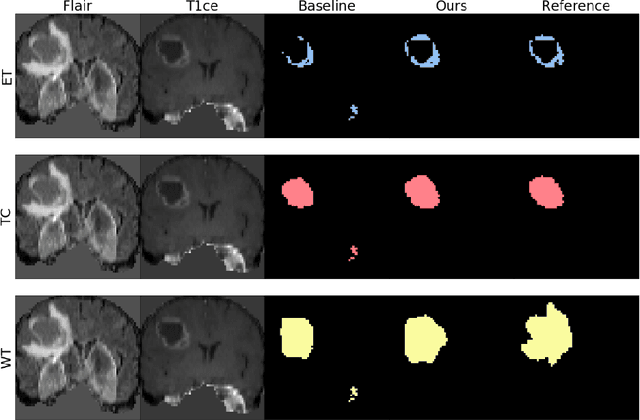
Abstract:The joint use of multiple imaging modalities for medical image segmentation has been widely studied in recent years. The fusion of information from different modalities has demonstrated to improve the segmentation accuracy, with respect to mono-modal segmentations, in several applications. However, acquiring multiple modalities is usually not possible in a clinical setting due to a limited number of physicians and scanners, and to limit costs and scan time. Most of the time, only one modality is acquired. In this paper, we propose KD-Net, a framework to transfer knowledge from a trained multi-modal network (teacher) to a mono-modal one (student). The proposed method is an adaptation of the generalized distillation framework where the student network is trained on a subset (1 modality) of the teacher's inputs (n modalities). We illustrate the effectiveness of the proposed framework in brain tumor segmentation with the BraTS 2018 dataset. Using different architectures, we show that the student network effectively learns from the teacher and always outperforms the baseline mono-modal network in terms of segmentation accuracy.
* MICCAI 2020
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge