Yihua Sun
Native Intelligence Emerges from Large-Scale Clinical Practice: A Retinal Foundation Model with Deployment Efficiency
Dec 16, 2025Abstract:Current retinal foundation models remain constrained by curated research datasets that lack authentic clinical context, and require extensive task-specific optimization for each application, limiting their deployment efficiency in low-resource settings. Here, we show that these barriers can be overcome by building clinical native intelligence directly from real-world medical practice. Our key insight is that large-scale telemedicine programs, where expert centers provide remote consultations across distributed facilities, represent a natural reservoir for learning clinical image interpretation. We present ReVision, a retinal foundation model that learns from the natural alignment between 485,980 color fundus photographs and their corresponding diagnostic reports, accumulated through a decade-long telemedicine program spanning 162 medical institutions across China. Through extensive evaluation across 27 ophthalmic benchmarks, we demonstrate that ReVison enables deployment efficiency with minimal local resources. Without any task-specific training, ReVision achieves zero-shot disease detection with an average AUROC of 0.946 across 12 public benchmarks and 0.952 on 3 independent clinical cohorts. When minimal adaptation is feasible, ReVision matches extensively fine-tuned alternatives while requiring orders of magnitude fewer trainable parameters and labeled examples. The learned representations also transfer effectively to new clinical sites, imaging domains, imaging modalities, and systemic health prediction tasks. In a prospective reader study with 33 ophthalmologists, ReVision's zero-shot assistance improved diagnostic accuracy by 14.8% across all experience levels. These results demonstrate that clinical native intelligence can be directly extracted from clinical archives without any further annotation to build medical AI systems suited to various low-resource settings.
LLM-RG4: Flexible and Factual Radiology Report Generation across Diverse Input Contexts
Dec 16, 2024Abstract:Drafting radiology reports is a complex task requiring flexibility, where radiologists tail content to available information and particular clinical demands. However, most current radiology report generation (RRG) models are constrained to a fixed task paradigm, such as predicting the full ``finding'' section from a single image, inherently involving a mismatch between inputs and outputs. The trained models lack the flexibility for diverse inputs and could generate harmful, input-agnostic hallucinations. To bridge the gap between current RRG models and the clinical demands in practice, we first develop a data generation pipeline to create a new MIMIC-RG4 dataset, which considers four common radiology report drafting scenarios and has perfectly corresponded input and output. Secondly, we propose a novel large language model (LLM) based RRG framework, namely LLM-RG4, which utilizes LLM's flexible instruction-following capabilities and extensive general knowledge. We further develop an adaptive token fusion module that offers flexibility to handle diverse scenarios with different input combinations, while minimizing the additional computational burden associated with increased input volumes. Besides, we propose a token-level loss weighting strategy to direct the model's attention towards positive and uncertain descriptions. Experimental results demonstrate that LLM-RG4 achieves state-of-the-art performance in both clinical efficiency and natural language generation on the MIMIC-RG4 and MIMIC-CXR datasets. We quantitatively demonstrate that our model has minimal input-agnostic hallucinations, whereas current open-source models commonly suffer from this problem.
Rib Suppression in Digital Chest Tomosynthesis
Mar 05, 2022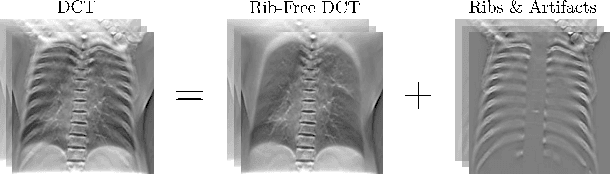
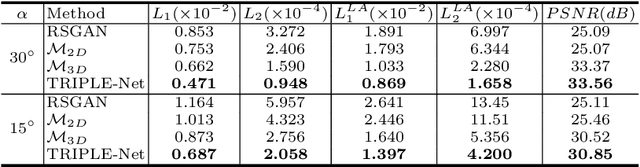
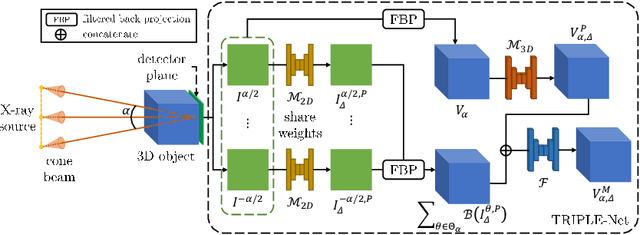
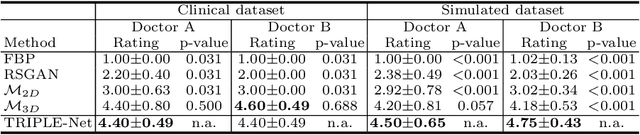
Abstract:Digital chest tomosynthesis (DCT) is a technique to produce sectional 3D images of a human chest for pulmonary disease screening, with 2D X-ray projections taken within an extremely limited range of angles. However, under the limited angle scenario, DCT contains strong artifacts caused by the presence of ribs, jamming the imaging quality of the lung area. Recently, great progress has been achieved for rib suppression in a single X-ray image, to reveal a clearer lung texture. We firstly extend the rib suppression problem to the 3D case at the software level. We propose a $\textbf{T}$omosynthesis $\textbf{RI}$b Su$\textbf{P}$pression and $\textbf{L}$ung $\textbf{E}$nhancement $\textbf{Net}$work (TRIPLE-Net) to model the 3D rib component and provide a rib-free DCT. TRIPLE-Net takes the advantages from both 2D and 3D domains, which model the ribs in DCT with the exact FBP procedure and 3D depth information, respectively. The experiments on simulated datasets and clinical data have shown the effectiveness of TRIPLE-Net to preserve lung details as well as improve the imaging quality of pulmonary diseases. Finally, an expert user study confirms our findings.
Relative distance matters for one-shot landmark detection
Mar 04, 2022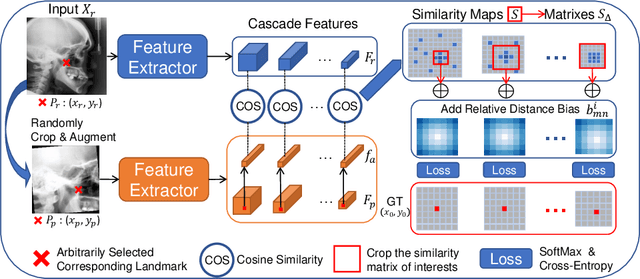
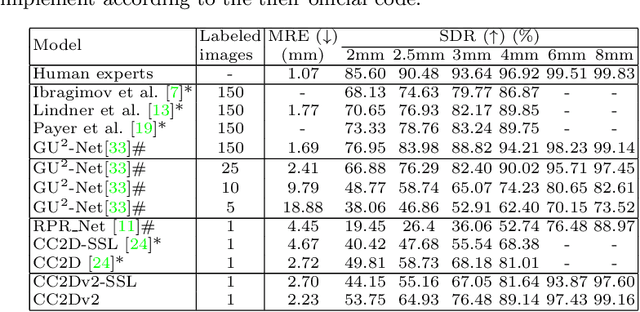
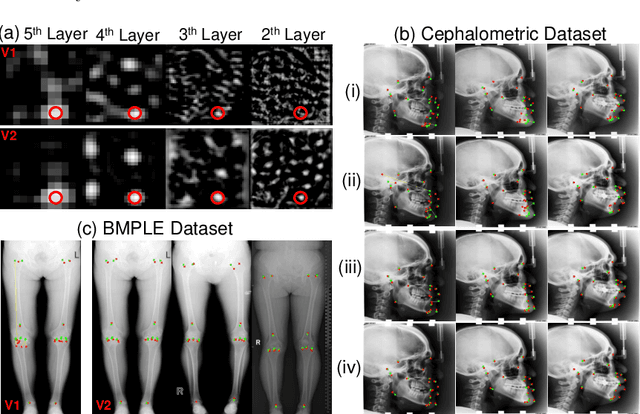
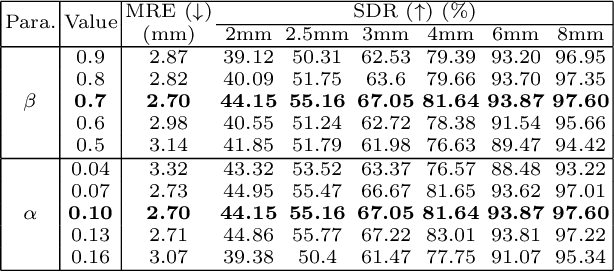
Abstract:Contrastive learning based methods such as cascade comparing to detect (CC2D) have shown great potential for one-shot medical landmark detection. However, the important cue of relative distance between landmarks is ignored in CC2D. In this paper, we upgrade CC2D to version II by incorporating a simple-yet-effective relative distance bias in the training stage, which is theoretically proved to encourage the encoder to project the relatively distant landmarks to the embeddings with low similarities. As consequence, CC2Dv2 is less possible to detect a wrong point far from the correct landmark. Furthermore, we present an open-source, landmark-labeled dataset for the measurement of biomechanical parameters of the lower extremity to alleviate the burden of orthopedic surgeons. The effectiveness of CC2Dv2 is evaluated on the public dataset from the ISBI 2015 Grand-Challenge of cephalometric radiographs and our new dataset, which greatly outperforms the state-of-the-art one-shot landmark detection approaches.
Review on Computer Vision in Gastric Cancer: Potential Efficient Tools for Diagnosis
May 31, 2020
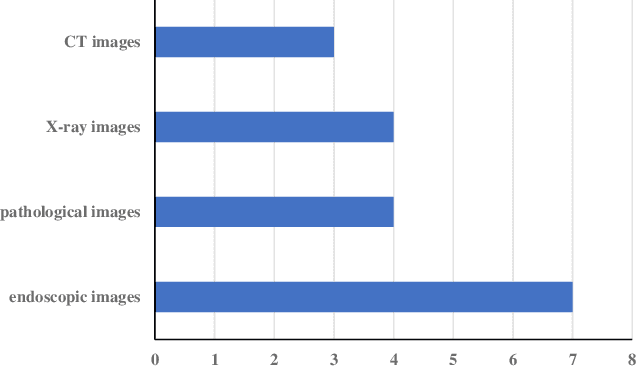
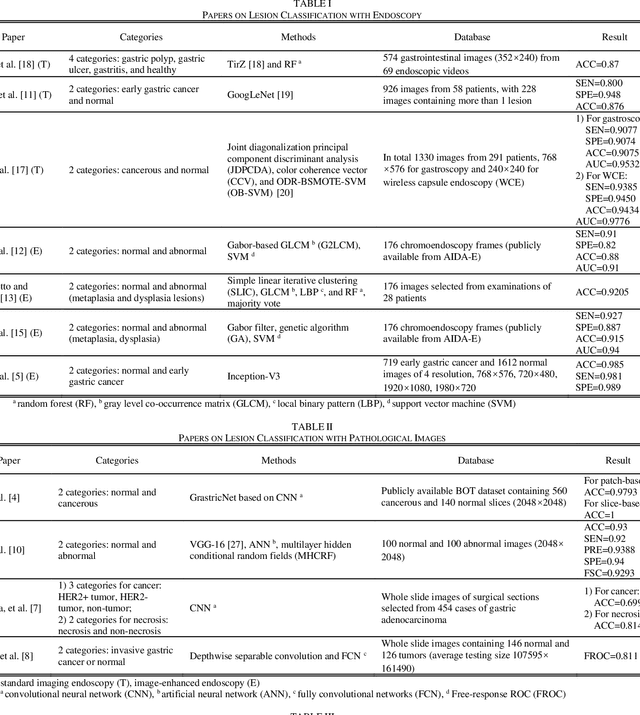
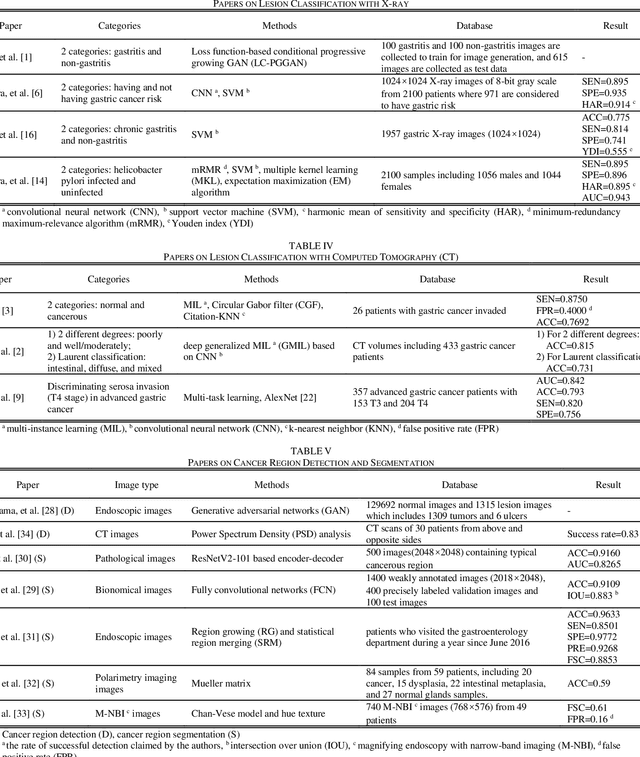
Abstract:Rapid diagnosis of gastric cancer is a great challenge for clinical doctors. Dramatic progress of computer vision on gastric cancer has been made recently and this review focuses on advances during the past five years. Different methods for data generation and augmentation are presented, and various approaches to extract discriminative features compared and evaluated. Classification and segmentation techniques are carefully discussed for assisting more precise diagnosis and timely treatment. For classification, various methods have been developed to better proceed specific images, such as images with rotation and estimated real-timely (endoscopy), high resolution images (histopathology), low diagnostic accuracy images (X-ray), poor contrast images of the soft-tissue with cavity (CT) or those images with insufficient annotation. For detection and segmentation, traditional methods and machine learning methods are compared. Application of those methods will greatly reduce the labor and time consumption for the diagnosis of gastric cancers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge