Heqin Zhu
U-Bench: A Comprehensive Understanding of U-Net through 100-Variant Benchmarking
Oct 08, 2025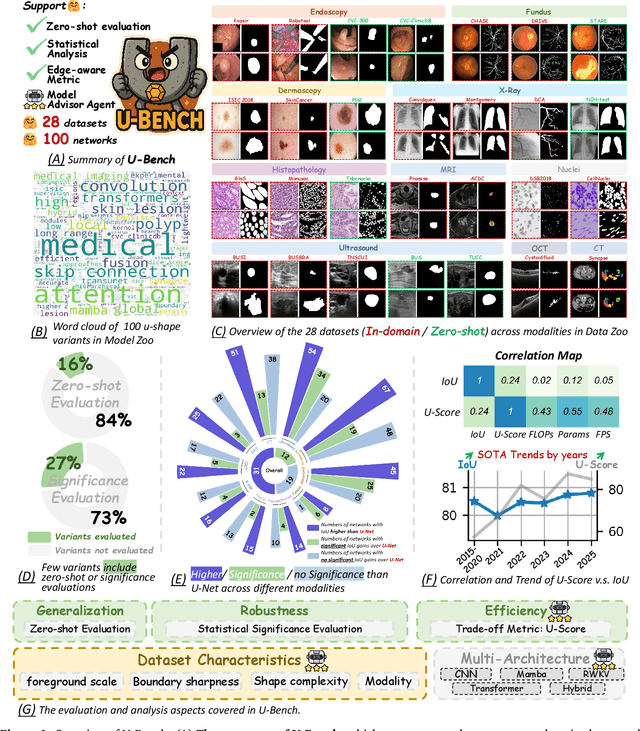


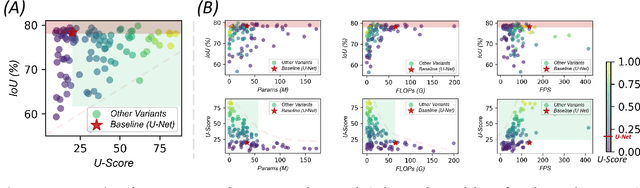
Abstract:Over the past decade, U-Net has been the dominant architecture in medical image segmentation, leading to the development of thousands of U-shaped variants. Despite its widespread adoption, there is still no comprehensive benchmark to systematically evaluate their performance and utility, largely because of insufficient statistical validation and limited consideration of efficiency and generalization across diverse datasets. To bridge this gap, we present U-Bench, the first large-scale, statistically rigorous benchmark that evaluates 100 U-Net variants across 28 datasets and 10 imaging modalities. Our contributions are threefold: (1) Comprehensive Evaluation: U-Bench evaluates models along three key dimensions: statistical robustness, zero-shot generalization, and computational efficiency. We introduce a novel metric, U-Score, which jointly captures the performance-efficiency trade-off, offering a deployment-oriented perspective on model progress. (2) Systematic Analysis and Model Selection Guidance: We summarize key findings from the large-scale evaluation and systematically analyze the impact of dataset characteristics and architectural paradigms on model performance. Based on these insights, we propose a model advisor agent to guide researchers in selecting the most suitable models for specific datasets and tasks. (3) Public Availability: We provide all code, models, protocols, and weights, enabling the community to reproduce our results and extend the benchmark with future methods. In summary, U-Bench not only exposes gaps in previous evaluations but also establishes a foundation for fair, reproducible, and practically relevant benchmarking in the next decade of U-Net-based segmentation models. The project can be accessed at: https://fenghetan9.github.io/ubench. Code is available at: https://github.com/FengheTan9/U-Bench.
HYATT-Net is Grand: A Hybrid Attention Network for Performant Anatomical Landmark Detection
Dec 16, 2024



Abstract:Anatomical landmark detection (ALD) from a medical image is crucial for a wide array of clinical applications. While existing methods achieve quite some success in ALD, they often struggle to balance global context with computational efficiency, particularly with high-resolution images, thereby leading to the rise of a natural question: where is the performance limit of ALD? In this paper, we aim to forge performant ALD by proposing a {\bf HY}brid {\bf ATT}ention {\bf Net}work (HYATT-Net) with the following designs: (i) A novel hybrid architecture that integrates CNNs and Transformers. Its core is the BiFormer module, utilizing Bi-Level Routing Attention for efficient attention to relevant image regions. This, combined with Attention Residual Module(ARM), enables precise local feature refinement guided by the global context. (ii) A Feature Fusion Correction Module that aggregates multi-scale features and thus mitigates a resolution loss. Deep supervision with a mean-square error loss on multi-resolution heatmaps optimizes the model. Experiments on five diverse datasets demonstrate state-of-the-art performance, surpassing existing methods in accuracy, robustness, and efficiency. The HYATT-Net provides a promising solution for accurate and efficient ALD in complex medical images. Our codes and data are already released at: \url{https://github.com/ECNUACRush/HYATT-Net}.
Hybrid Attention Network: An efficient approach for anatomy-free landmark detection
Dec 09, 2024



Abstract:Accurate anatomical landmark detection in medical images is crucial for clinical applications. Existing methods often struggle to balance global context with computational efficiency, particularly with high-resolution images. This paper introduces the Hybrid Attention Network(HAN), a novel hybrid architecture integrating CNNs and Transformers. Its core is the BiFormer module, utilizing Bi-Level Routing Attention (BRA) for efficient attention to relevant image regions. This, combined with Convolutional Attention Blocks (CAB) enhanced by CBAM, enables precise local feature refinement guided by the global context. A Feature Fusion Correction Module (FFCM) integrates multi-scale features, mitigating resolution loss. Deep supervision with MSE loss on multi-resolution heatmaps optimizes the model. Experiments on five diverse datasets demonstrate state-of-the-art performance, surpassing existing methods in accuracy, robustness, and efficiency. The HAN provides a promising solution for accurate and efficient anatomical landmark detection in complex medical images. Our codes and data will be released soon at: \url{https://github.com/MIRACLE-Center/}.
HySparK: Hybrid Sparse Masking for Large Scale Medical Image Pre-Training
Aug 11, 2024



Abstract:The generative self-supervised learning strategy exhibits remarkable learning representational capabilities. However, there is limited attention to end-to-end pre-training methods based on a hybrid architecture of CNN and Transformer, which can learn strong local and global representations simultaneously. To address this issue, we propose a generative pre-training strategy called Hybrid Sparse masKing (HySparK) based on masked image modeling and apply it to large-scale pre-training on medical images. First, we perform a bottom-up 3D hybrid masking strategy on the encoder to keep consistency masking. Then we utilize sparse convolution for the top CNNs and encode unmasked patches for the bottom vision Transformers. Second, we employ a simple hierarchical decoder with skip-connections to achieve dense multi-scale feature reconstruction. Third, we implement our pre-training method on a collection of multiple large-scale 3D medical imaging datasets. Extensive experiments indicate that our proposed pre-training strategy demonstrates robust transfer-ability in supervised downstream tasks and sheds light on HySparK's promising prospects. The code is available at https://github.com/FengheTan9/HySparK
Slide-SAM: Medical SAM Meets Sliding Window
Dec 05, 2023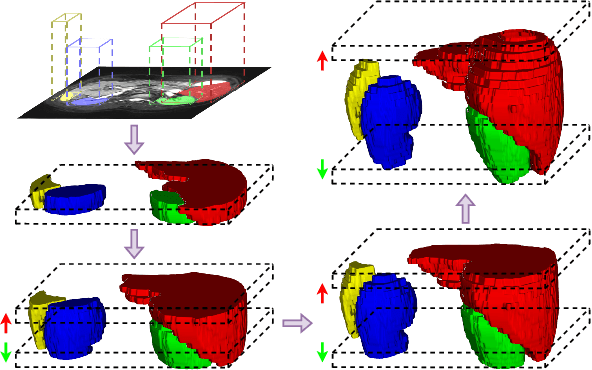
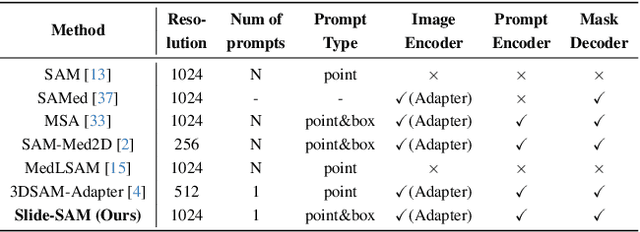
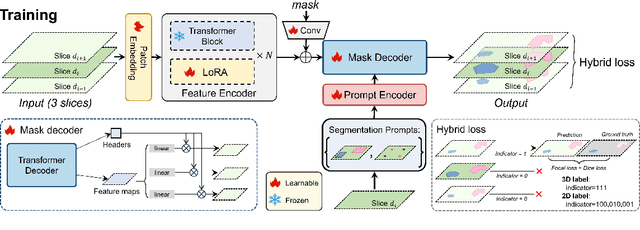
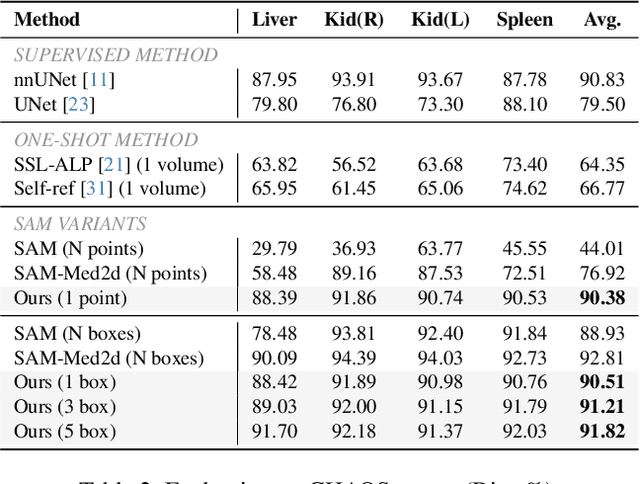
Abstract:The Segment Anything Model (SAM) has achieved a notable success in two-dimensional image segmentation in natural images. However, the substantial gap between medical and natural images hinders its direct application to medical image segmentation tasks. Particularly in 3D medical images, SAM struggles to learn contextual relationships between slices, limiting its practical applicability. Moreover, applying 2D SAM to 3D images requires prompting the entire volume, which is time- and label-consuming. To address these problems, we propose Slide-SAM, which treats a stack of three adjacent slices as a prediction window. It firstly takes three slices from a 3D volume and point- or bounding box prompts on the central slice as inputs to predict segmentation masks for all three slices. Subsequently, the masks of the top and bottom slices are then used to generate new prompts for adjacent slices. Finally, step-wise prediction can be achieved by sliding the prediction window forward or backward through the entire volume. Our model is trained on multiple public and private medical datasets and demonstrates its effectiveness through extensive 3D segmetnation experiments, with the help of minimal prompts. Code is available at \url{https://github.com/Curli-quan/Slide-SAM}.
UOD: Universal One-shot Detection of Anatomical Landmarks
Jun 14, 2023Abstract:One-shot medical landmark detection gains much attention and achieves great success for its label-efficient training process. However, existing one-shot learning methods are highly specialized in a single domain and suffer domain preference heavily in the situation of multi-domain unlabeled data. Moreover, one-shot learning is not robust that it faces performance drop when annotating a sub-optimal image. To tackle these issues, we resort to developing a domain-adaptive one-shot landmark detection framework for handling multi-domain medical images, named Universal One-shot Detection (UOD). UOD consists of two stages and two corresponding universal models which are designed as combinations of domain-specific modules and domain-shared modules. In the first stage, a domain-adaptive convolution model is self-supervised learned to generate pseudo landmark labels. In the second stage, we design a domain-adaptive transformer to eliminate domain preference and build the global context for multi-domain data. Even though only one annotated sample from each domain is available for training, the domain-shared modules help UOD aggregate all one-shot samples to detect more robust and accurate landmarks. We investigated both qualitatively and quantitatively the proposed UOD on three widely-used public X-ray datasets in different anatomical domains (i.e., head, hand, chest) and obtained state-of-the-art performances in each domain.
Unsupervised augmentation optimization for few-shot medical image segmentation
Jun 08, 2023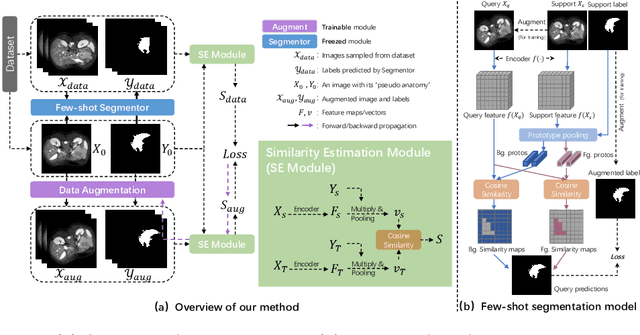
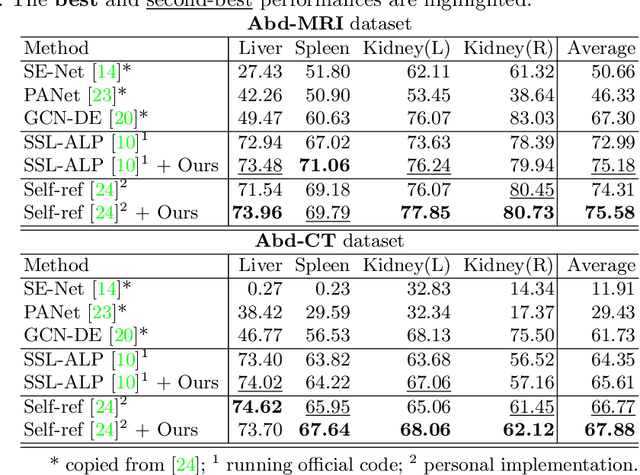
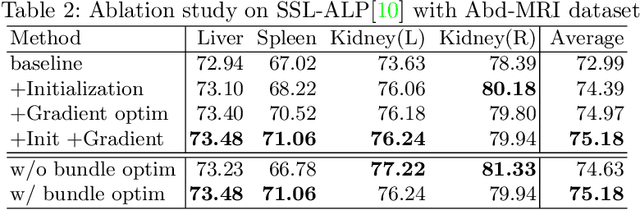
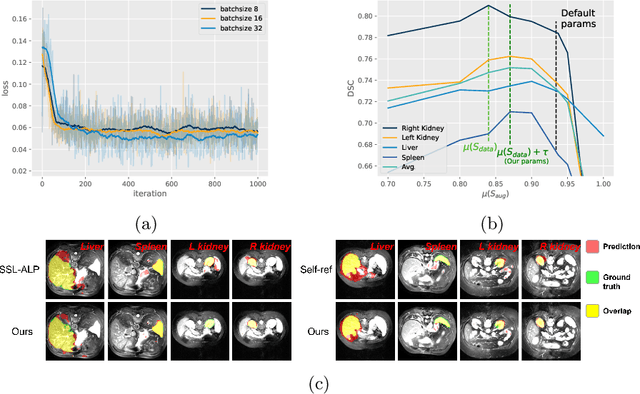
Abstract:The augmentation parameters matter to few-shot semantic segmentation since they directly affect the training outcome by feeding the networks with varying perturbated samples. However, searching optimal augmentation parameters for few-shot segmentation models without annotations is a challenge that current methods fail to address. In this paper, we first propose a framework to determine the ``optimal'' parameters without human annotations by solving a distribution-matching problem between the intra-instance and intra-class similarity distribution, with the intra-instance similarity describing the similarity between the original sample of a particular anatomy and its augmented ones and the intra-class similarity representing the similarity between the selected sample and the others in the same class. Extensive experiments demonstrate the superiority of our optimized augmentation in boosting few-shot segmentation models. We greatly improve the top competing method by 1.27\% and 1.11\% on Abd-MRI and Abd-CT datasets, respectively, and even achieve a significant improvement for SSL-ALP on the left kidney by 3.39\% on the Abd-CT dataset.
PELE scores: Pelvic X-ray Landmark Detection by Pelvis Extraction and Enhancement
May 07, 2023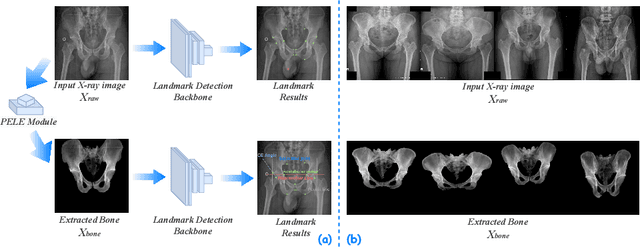

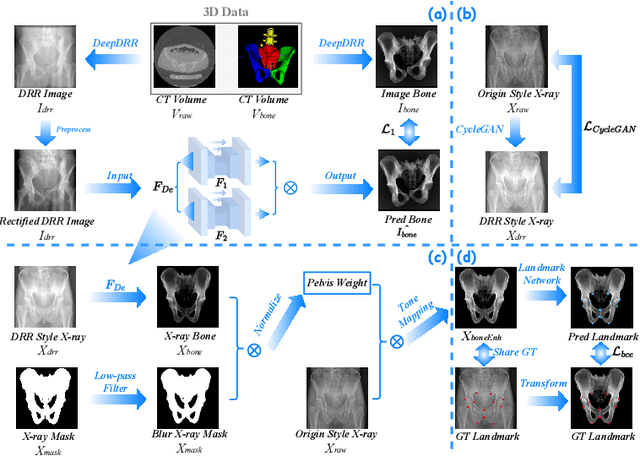

Abstract:The pelvis, the lower part of the trunk, supports and balances the trunk. Landmark detection from a pelvic X-ray (PXR) facilitates downstream analysis and computer-assisted diagnosis and treatment of pelvic diseases. Although PXRs have the advantages of low radiation and reduced cost compared to computed tomography (CT) images, their 2D pelvis-tissue superposition of 3D structures confuses clinical decision-making. In this paper, we propose a PELvis Extraction (PELE) module that utilizes 3D prior anatomical knowledge in CT to guide and well isolate the pelvis from PXRs, thereby eliminating the influence of soft tissue. We conduct an extensive evaluation based on two public datasets and one private dataset, totaling 850 PXRs. The experimental results show that the proposed PELE module significantly improves the accuracy of PXRs landmark detection and achieves state-of-the-art performances in several benchmark metrics, thus better serving downstream tasks.
DFTR: Depth-supervised Fusion Transformer for Salient Object Detection
Apr 11, 2022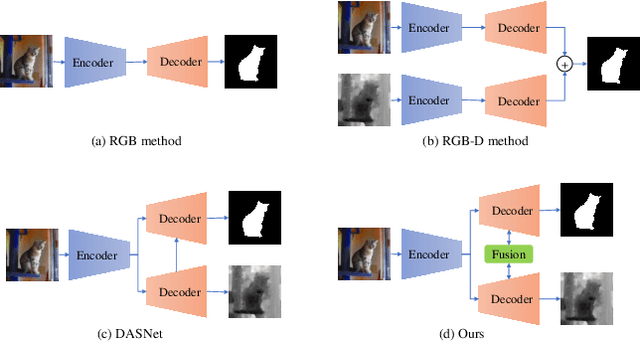
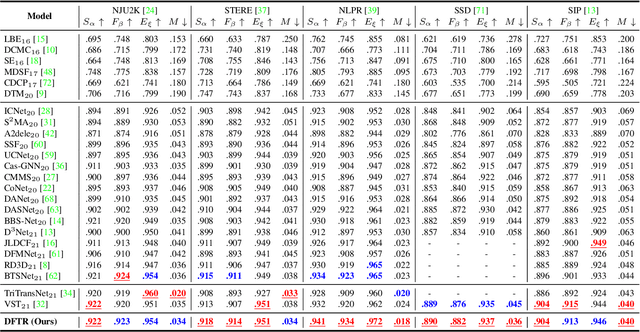
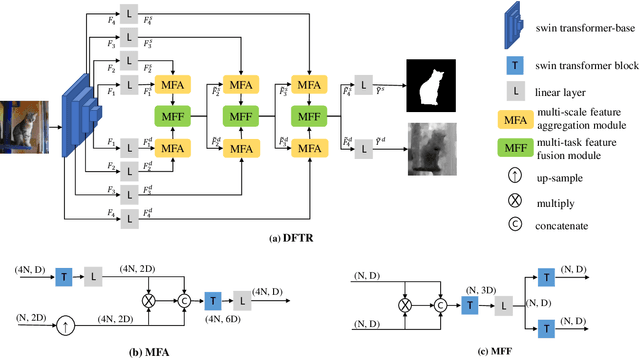
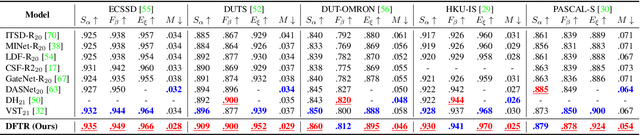
Abstract:Automated salient object detection (SOD) plays an increasingly crucial role in many computer vision applications. By reformulating the depth information as supervision rather than as input, depth-supervised convolutional neural networks (CNN) have achieved promising results on both RGB and RGB-D SOD scenarios with the merits of no requirements for extra depth networks and depth inputs in the inference stage. This paper, for the first time, seeks to expand the applicability of depth supervision to the Transformer architecture. Specifically, we develop a Depth-supervised Fusion TRansformer (DFTR), to further improve the accuracy of both RGB and RGB-D SOD. The proposed DFTR involves three primary features: 1) DFTR, to the best of our knowledge, is the first pure Transformer-based model for depth-supervised SOD; 2) A multi-scale feature aggregation (MFA) module is proposed to fully exploit the multi-scale features encoded by the Swin Transformer in a coarse-to-fine manner; 3) To enable bidirectional information flow across different streams of features, a novel multi-stage feature fusion (MFF) module is further integrated into our DFTR with the emphasis on salient regions at different network learning stages. We extensively evaluate the proposed DFTR on ten benchmarking datasets. Experimental results show that our DFTR consistently outperforms the existing state-of-the-art methods for both RGB and RGB-D SOD tasks. The code and model will be made publicly available.
DATR: Domain-adaptive transformer for multi-domain landmark detection
Mar 12, 2022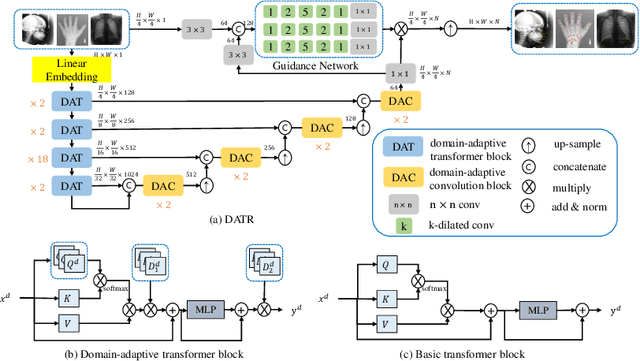
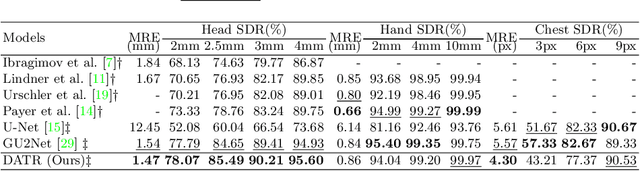
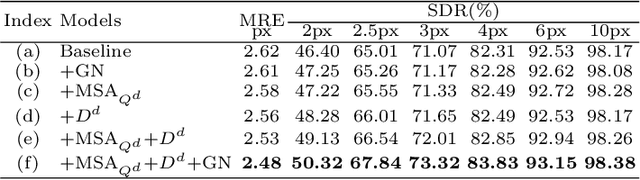
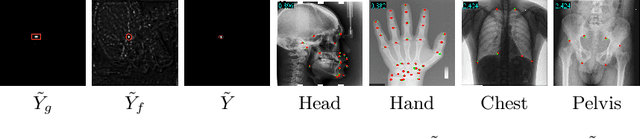
Abstract:Accurate anatomical landmark detection plays an increasingly vital role in medical image analysis. Although existing methods achieve satisfying performance, they are mostly based on CNN and specialized for a single domain say associated with a particular anatomical region. In this work, we propose a universal model for multi-domain landmark detection by taking advantage of transformer for modeling long dependencies and develop a domain-adaptive transformer model, named as DATR, which is trained on multiple mixed datasets from different anatomies and capable of detecting landmarks of any image from those anatomies. The proposed DATR exhibits three primary features: (i) It is the first universal model which introduces transformer as an encoder for multi-anatomy landmark detection; (ii) We design a domain-adaptive transformer for anatomy-aware landmark detection, which can be effectively extended to any other transformer network; (iii) Following previous studies, we employ a light-weighted guidance network, which encourages the transformer network to detect more accurate landmarks. We carry out experiments on three widely used X-ray datasets for landmark detection, which have 1,588 images and 62 landmarks in total, including three different anatomies (head, hand, and chest). Experimental results demonstrate that our proposed DATR achieves state-of-the-art performances by most metrics and behaves much better than any previous convolution-based models. The code will be released publicly.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge