Fenghe Tang
UCAD: Uncertainty-guided Contour-aware Displacement for semi-supervised medical image segmentation
Jan 24, 2026Abstract:Existing displacement strategies in semi-supervised segmentation only operate on rectangular regions, ignoring anatomical structures and resulting in boundary distortions and semantic inconsistency. To address these issues, we propose UCAD, an Uncertainty-Guided Contour-Aware Displacement framework for semi-supervised medical image segmentation that preserves contour-aware semantics while enhancing consistency learning. Our UCAD leverages superpixels to generate anatomically coherent regions aligned with anatomy boundaries, and an uncertainty-guided selection mechanism to selectively displace challenging regions for better consistency learning. We further propose a dynamic uncertainty-weighted consistency loss, which adaptively stabilizes training and effectively regularizes the model on unlabeled regions. Extensive experiments demonstrate that UCAD consistently outperforms state-of-the-art semi-supervised segmentation methods, achieving superior segmentation accuracy under limited annotation. The code is available at:https://github.com/dcb937/UCAD.
Equivariant Sampling for Improving Diffusion Model-based Image Restoration
Nov 13, 2025

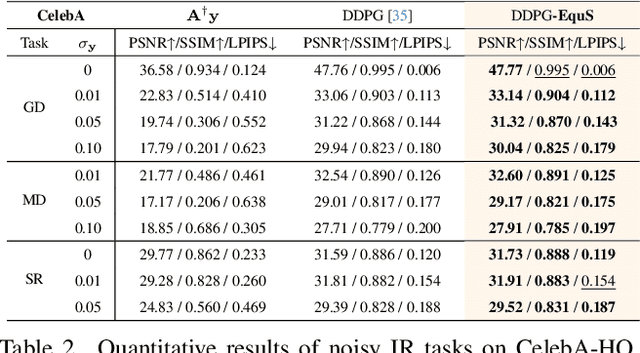

Abstract:Recent advances in generative models, especially diffusion models, have significantly improved image restoration (IR) performance. However, existing problem-agnostic diffusion model-based image restoration (DMIR) methods face challenges in fully leveraging diffusion priors, resulting in suboptimal performance. In this paper, we address the limitations of current problem-agnostic DMIR methods by analyzing their sampling process and providing effective solutions. We introduce EquS, a DMIR method that imposes equivariant information through dual sampling trajectories. To further boost EquS, we propose the Timestep-Aware Schedule (TAS) and introduce EquS$^+$. TAS prioritizes deterministic steps to enhance certainty and sampling efficiency. Extensive experiments on benchmarks demonstrate that our method is compatible with previous problem-agnostic DMIR methods and significantly boosts their performance without increasing computational costs. Our code is available at https://github.com/FouierL/EquS.
U-Bench: A Comprehensive Understanding of U-Net through 100-Variant Benchmarking
Oct 08, 2025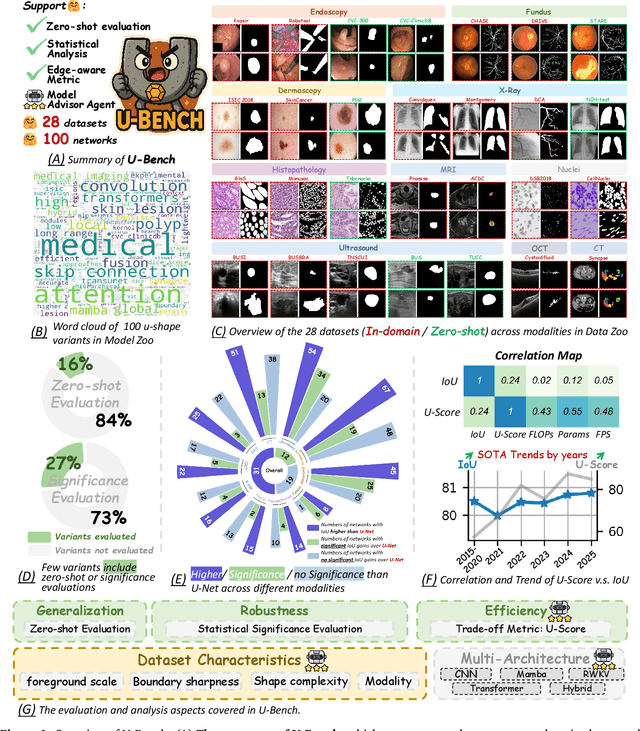


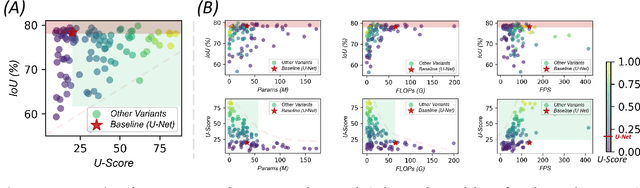
Abstract:Over the past decade, U-Net has been the dominant architecture in medical image segmentation, leading to the development of thousands of U-shaped variants. Despite its widespread adoption, there is still no comprehensive benchmark to systematically evaluate their performance and utility, largely because of insufficient statistical validation and limited consideration of efficiency and generalization across diverse datasets. To bridge this gap, we present U-Bench, the first large-scale, statistically rigorous benchmark that evaluates 100 U-Net variants across 28 datasets and 10 imaging modalities. Our contributions are threefold: (1) Comprehensive Evaluation: U-Bench evaluates models along three key dimensions: statistical robustness, zero-shot generalization, and computational efficiency. We introduce a novel metric, U-Score, which jointly captures the performance-efficiency trade-off, offering a deployment-oriented perspective on model progress. (2) Systematic Analysis and Model Selection Guidance: We summarize key findings from the large-scale evaluation and systematically analyze the impact of dataset characteristics and architectural paradigms on model performance. Based on these insights, we propose a model advisor agent to guide researchers in selecting the most suitable models for specific datasets and tasks. (3) Public Availability: We provide all code, models, protocols, and weights, enabling the community to reproduce our results and extend the benchmark with future methods. In summary, U-Bench not only exposes gaps in previous evaluations but also establishes a foundation for fair, reproducible, and practically relevant benchmarking in the next decade of U-Net-based segmentation models. The project can be accessed at: https://fenghetan9.github.io/ubench. Code is available at: https://github.com/FengheTan9/U-Bench.
SimCroP: Radiograph Representation Learning with Similarity-driven Cross-granularity Pre-training
Sep 10, 2025Abstract:Medical vision-language pre-training shows great potential in learning representative features from massive paired radiographs and reports. However, in computed tomography (CT) scans, the distribution of lesions which contain intricate structures is characterized by spatial sparsity. Besides, the complex and implicit relationships between different pathological descriptions in each sentence of the report and their corresponding sub-regions in radiographs pose additional challenges. In this paper, we propose a Similarity-Driven Cross-Granularity Pre-training (SimCroP) framework on chest CTs, which combines similarity-driven alignment and cross-granularity fusion to improve radiograph interpretation. We first leverage multi-modal masked modeling to optimize the encoder for understanding precise low-level semantics from radiographs. Then, similarity-driven alignment is designed to pre-train the encoder to adaptively select and align the correct patches corresponding to each sentence in reports. The cross-granularity fusion module integrates multimodal information across instance level and word-patch level, which helps the model better capture key pathology structures in sparse radiographs, resulting in improved performance for multi-scale downstream tasks. SimCroP is pre-trained on a large-scale paired CT-reports dataset and validated on image classification and segmentation tasks across five public datasets. Experimental results demonstrate that SimCroP outperforms both cutting-edge medical self-supervised learning methods and medical vision-language pre-training methods. Codes and models are available at https://github.com/ToniChopp/SimCroP.
U-RWKV: Lightweight medical image segmentation with direction-adaptive RWKV
Jul 15, 2025Abstract:Achieving equity in healthcare accessibility requires lightweight yet high-performance solutions for medical image segmentation, particularly in resource-limited settings. Existing methods like U-Net and its variants often suffer from limited global Effective Receptive Fields (ERFs), hindering their ability to capture long-range dependencies. To address this, we propose U-RWKV, a novel framework leveraging the Recurrent Weighted Key-Value(RWKV) architecture, which achieves efficient long-range modeling at O(N) computational cost. The framework introduces two key innovations: the Direction-Adaptive RWKV Module(DARM) and the Stage-Adaptive Squeeze-and-Excitation Module(SASE). DARM employs Dual-RWKV and QuadScan mechanisms to aggregate contextual cues across images, mitigating directional bias while preserving global context and maintaining high computational efficiency. SASE dynamically adapts its architecture to different feature extraction stages, balancing high-resolution detail preservation and semantic relationship capture. Experiments demonstrate that U-RWKV achieves state-of-the-art segmentation performance with high computational efficiency, offering a practical solution for democratizing advanced medical imaging technologies in resource-constrained environments. The code is available at https://github.com/hbyecoding/U-RWKV.
AA-CLIP: Enhancing Zero-shot Anomaly Detection via Anomaly-Aware CLIP
Mar 09, 2025Abstract:Anomaly detection (AD) identifies outliers for applications like defect and lesion detection. While CLIP shows promise for zero-shot AD tasks due to its strong generalization capabilities, its inherent Anomaly-Unawareness leads to limited discrimination between normal and abnormal features. To address this problem, we propose Anomaly-Aware CLIP (AA-CLIP), which enhances CLIP's anomaly discrimination ability in both text and visual spaces while preserving its generalization capability. AA-CLIP is achieved through a straightforward yet effective two-stage approach: it first creates anomaly-aware text anchors to differentiate normal and abnormal semantics clearly, then aligns patch-level visual features with these anchors for precise anomaly localization. This two-stage strategy, with the help of residual adapters, gradually adapts CLIP in a controlled manner, achieving effective AD while maintaining CLIP's class knowledge. Extensive experiments validate AA-CLIP as a resource-efficient solution for zero-shot AD tasks, achieving state-of-the-art results in industrial and medical applications. The code is available at https://github.com/Mwxinnn/AA-CLIP.
* 8 pages, 7 figures
Hi-End-MAE: Hierarchical encoder-driven masked autoencoders are stronger vision learners for medical image segmentation
Feb 12, 2025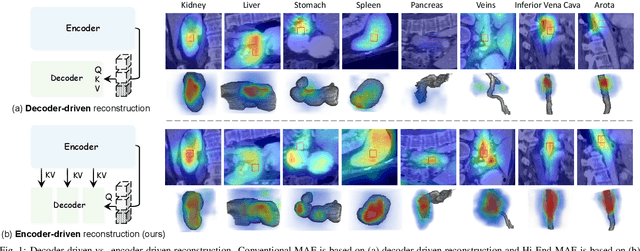
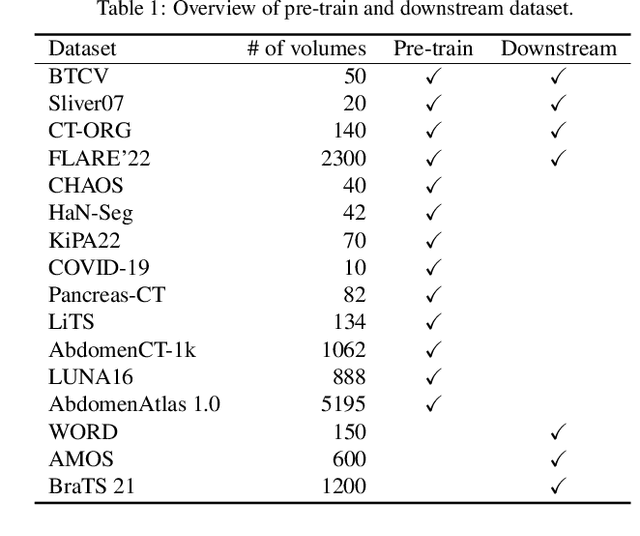
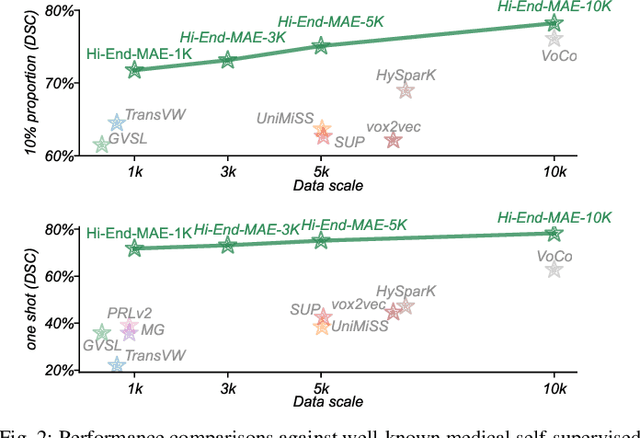
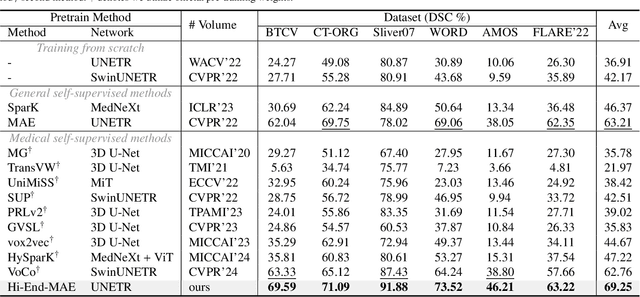
Abstract:Medical image segmentation remains a formidable challenge due to the label scarcity. Pre-training Vision Transformer (ViT) through masked image modeling (MIM) on large-scale unlabeled medical datasets presents a promising solution, providing both computational efficiency and model generalization for various downstream tasks. However, current ViT-based MIM pre-training frameworks predominantly emphasize local aggregation representations in output layers and fail to exploit the rich representations across different ViT layers that better capture fine-grained semantic information needed for more precise medical downstream tasks. To fill the above gap, we hereby present Hierarchical Encoder-driven MAE (Hi-End-MAE), a simple yet effective ViT-based pre-training solution, which centers on two key innovations: (1) Encoder-driven reconstruction, which encourages the encoder to learn more informative features to guide the reconstruction of masked patches; and (2) Hierarchical dense decoding, which implements a hierarchical decoding structure to capture rich representations across different layers. We pre-train Hi-End-MAE on a large-scale dataset of 10K CT scans and evaluated its performance across seven public medical image segmentation benchmarks. Extensive experiments demonstrate that Hi-End-MAE achieves superior transfer learning capabilities across various downstream tasks, revealing the potential of ViT in medical imaging applications. The code is available at: https://github.com/FengheTan9/Hi-End-MAE
3DGR-CAR: Coronary artery reconstruction from ultra-sparse 2D X-ray views with a 3D Gaussians representation
Oct 01, 2024Abstract:Reconstructing 3D coronary arteries is important for coronary artery disease diagnosis, treatment planning and operation navigation. Traditional reconstruction techniques often require many projections, while reconstruction from sparse-view X-ray projections is a potential way of reducing radiation dose. However, the extreme sparsity of coronary arteries in a 3D volume and ultra-limited number of projections pose significant challenges for efficient and accurate 3D reconstruction. To this end, we propose 3DGR-CAR, a 3D Gaussian Representation for Coronary Artery Reconstruction from ultra-sparse X-ray projections. We leverage 3D Gaussian representation to avoid the inefficiency caused by the extreme sparsity of coronary artery data and propose a Gaussian center predictor to overcome the noisy Gaussian initialization from ultra-sparse view projections. The proposed scheme enables fast and accurate 3D coronary artery reconstruction with only 2 views. Experimental results on two datasets indicate that the proposed approach significantly outperforms other methods in terms of voxel accuracy and visual quality of coronary arteries. The code will be available in https://github.com/windrise/3DGR-CAR.
MambaMIM: Pre-training Mamba with State Space Token-interpolation
Aug 15, 2024Abstract:Generative self-supervised learning demonstrates outstanding representation learning capabilities in both Convolutional Neural Networks (CNNs) and Vision Transformers (ViTs). However, there are currently no generative pre-training methods related to selective state space models (Mamba) that can handle long-range dependencies effectively. To address this challenge, we introduce a generative self-supervised learning method for Mamba (MambaMIM) based on Selective Structure State Space Sequence Token-interpolation (S6T), a general-purpose pre-training method for arbitrary Mamba architectures. Our method, MambaMIM, incorporates a bottom-up 3D hybrid masking strategy in the encoder to maintain masking consistency across different architectures. Additionally, S6T is employed to learn causal relationships between the masked sequence in the state space. MambaMIM can be used on any single or hybrid Mamba architectures to enhance the Mamba long-range representation capability. Extensive downstream experiments reveal the feasibility and advancement of using Mamba for pre-training medical image tasks. The code is available at: https://github.com/FengheTan9/MambaMIM
HySparK: Hybrid Sparse Masking for Large Scale Medical Image Pre-Training
Aug 11, 2024



Abstract:The generative self-supervised learning strategy exhibits remarkable learning representational capabilities. However, there is limited attention to end-to-end pre-training methods based on a hybrid architecture of CNN and Transformer, which can learn strong local and global representations simultaneously. To address this issue, we propose a generative pre-training strategy called Hybrid Sparse masKing (HySparK) based on masked image modeling and apply it to large-scale pre-training on medical images. First, we perform a bottom-up 3D hybrid masking strategy on the encoder to keep consistency masking. Then we utilize sparse convolution for the top CNNs and encode unmasked patches for the bottom vision Transformers. Second, we employ a simple hierarchical decoder with skip-connections to achieve dense multi-scale feature reconstruction. Third, we implement our pre-training method on a collection of multiple large-scale 3D medical imaging datasets. Extensive experiments indicate that our proposed pre-training strategy demonstrates robust transfer-ability in supervised downstream tasks and sheds light on HySparK's promising prospects. The code is available at https://github.com/FengheTan9/HySparK
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge