Ronghao Xu
H3DE-Net: Efficient and Accurate 3D Landmark Detection in Medical Imaging
Feb 20, 2025Abstract:3D landmark detection is a critical task in medical image analysis, and accurately detecting anatomical landmarks is essential for subsequent medical imaging tasks. However, mainstream deep learning methods in this field struggle to simultaneously capture fine-grained local features and model global spatial relationships, while maintaining a balance between accuracy and computational efficiency. Local feature extraction requires capturing fine-grained anatomical details, while global modeling requires understanding the spatial relationships within complex anatomical structures. The high-dimensional nature of 3D volume further exacerbates these challenges, as landmarks are sparsely distributed, leading to significant computational costs. Therefore, achieving efficient and precise 3D landmark detection remains a pressing challenge in medical image analysis. In this work, We propose a \textbf{H}ybrid \textbf{3}D \textbf{DE}tection \textbf{Net}(H3DE-Net), a novel framework that combines CNNs for local feature extraction with a lightweight attention mechanism designed to efficiently capture global dependencies in 3D volumetric data. This mechanism employs a hierarchical routing strategy to reduce computational cost while maintaining global context modeling. To our knowledge, H3DE-Net is the first 3D landmark detection model that integrates such a lightweight attention mechanism with CNNs. Additionally, integrating multi-scale feature fusion further enhances detection accuracy and robustness. Experimental results on a public CT dataset demonstrate that H3DE-Net achieves state-of-the-art(SOTA) performance, significantly improving accuracy and robustness, particularly in scenarios with missing landmarks or complex anatomical variations. We aready open-source our project, including code, data and model weights.
HySparK: Hybrid Sparse Masking for Large Scale Medical Image Pre-Training
Aug 11, 2024



Abstract:The generative self-supervised learning strategy exhibits remarkable learning representational capabilities. However, there is limited attention to end-to-end pre-training methods based on a hybrid architecture of CNN and Transformer, which can learn strong local and global representations simultaneously. To address this issue, we propose a generative pre-training strategy called Hybrid Sparse masKing (HySparK) based on masked image modeling and apply it to large-scale pre-training on medical images. First, we perform a bottom-up 3D hybrid masking strategy on the encoder to keep consistency masking. Then we utilize sparse convolution for the top CNNs and encode unmasked patches for the bottom vision Transformers. Second, we employ a simple hierarchical decoder with skip-connections to achieve dense multi-scale feature reconstruction. Third, we implement our pre-training method on a collection of multiple large-scale 3D medical imaging datasets. Extensive experiments indicate that our proposed pre-training strategy demonstrates robust transfer-ability in supervised downstream tasks and sheds light on HySparK's promising prospects. The code is available at https://github.com/FengheTan9/HySparK
ForgeryNet -- Face Forgery Analysis Challenge 2021: Methods and Results
Dec 15, 2021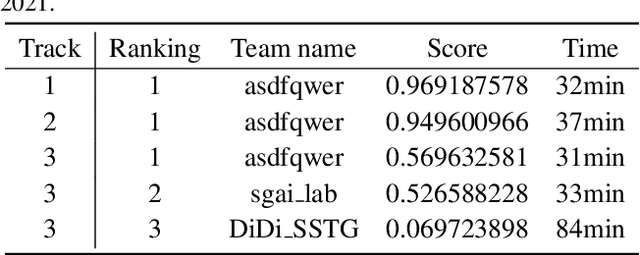
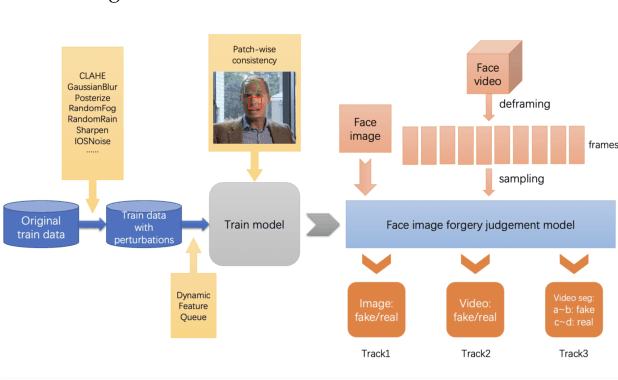
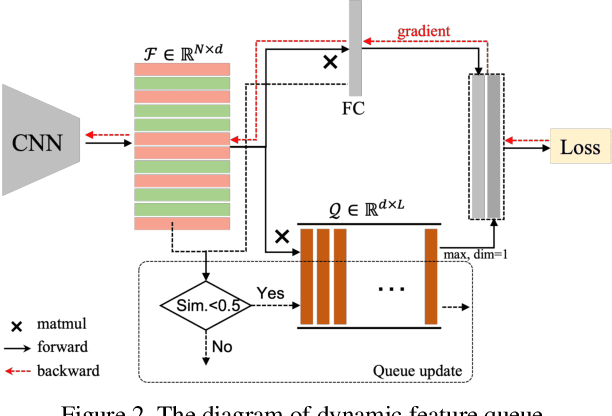
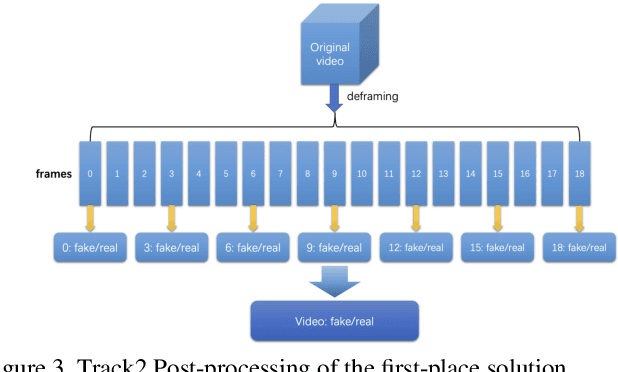
Abstract:The rapid progress of photorealistic synthesis techniques has reached a critical point where the boundary between real and manipulated images starts to blur. Recently, a mega-scale deep face forgery dataset, ForgeryNet which comprised of 2.9 million images and 221,247 videos has been released. It is by far the largest publicly available in terms of data-scale, manipulations (7 image-level approaches, 8 video-level approaches), perturbations (36 independent and more mixed perturbations), and annotations (6.3 million classification labels, 2.9 million manipulated area annotations, and 221,247 temporal forgery segment labels). This paper reports methods and results in the ForgeryNet - Face Forgery Analysis Challenge 2021, which employs the ForgeryNet benchmark. The model evaluation is conducted offline on the private test set. A total of 186 participants registered for the competition, and 11 teams made valid submissions. We will analyze the top-ranked solutions and present some discussion on future work directions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge