Chenxu Wu
Adapting Reinforcement Learning for Path Planning in Constrained Parking Scenarios
Jan 30, 2026Abstract:Real-time path planning in constrained environments remains a fundamental challenge for autonomous systems. Traditional classical planners, while effective under perfect perception assumptions, are often sensitive to real-world perception constraints and rely on online search procedures that incur high computational costs. In complex surroundings, this renders real-time deployment prohibitive. To overcome these limitations, we introduce a Deep Reinforcement Learning (DRL) framework for real-time path planning in parking scenarios. In particular, we focus on challenging scenes with tight spaces that require a high number of reversal maneuvers and adjustments. Unlike classical planners, our solution does not require ideal and structured perception, and in principle, could avoid the need for additional modules such as localization and tracking, resulting in a simpler and more practical implementation. Also, at test time, the policy generates actions through a single forward pass at each step, which is lightweight enough for real-time deployment. The task is formulated as a sequential decision-making problem grounded in a bicycle model dynamics, enabling the agent to directly learn navigation policies that respect vehicle kinematics and environmental constraints in the closed-loop setting. A new benchmark is developed to support both training and evaluation, capturing diverse and challenging scenarios. Our approach achieves state-of-the-art success rates and efficiency, surpassing classical planner baselines by +96% in success rate and +52% in efficiency. Furthermore, we release our benchmark as an open-source resource for the community to foster future research in autonomous systems. The benchmark and accompanying tools are available at https://github.com/dqm5rtfg9b-collab/Constrained_Parking_Scenarios.
Equivariant Sampling for Improving Diffusion Model-based Image Restoration
Nov 13, 2025

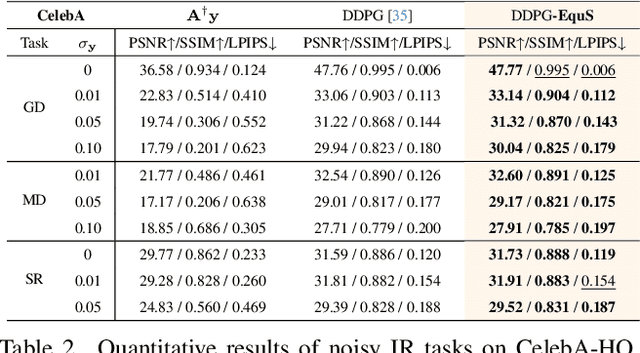

Abstract:Recent advances in generative models, especially diffusion models, have significantly improved image restoration (IR) performance. However, existing problem-agnostic diffusion model-based image restoration (DMIR) methods face challenges in fully leveraging diffusion priors, resulting in suboptimal performance. In this paper, we address the limitations of current problem-agnostic DMIR methods by analyzing their sampling process and providing effective solutions. We introduce EquS, a DMIR method that imposes equivariant information through dual sampling trajectories. To further boost EquS, we propose the Timestep-Aware Schedule (TAS) and introduce EquS$^+$. TAS prioritizes deterministic steps to enhance certainty and sampling efficiency. Extensive experiments on benchmarks demonstrate that our method is compatible with previous problem-agnostic DMIR methods and significantly boosts their performance without increasing computational costs. Our code is available at https://github.com/FouierL/EquS.
U-Bench: A Comprehensive Understanding of U-Net through 100-Variant Benchmarking
Oct 08, 2025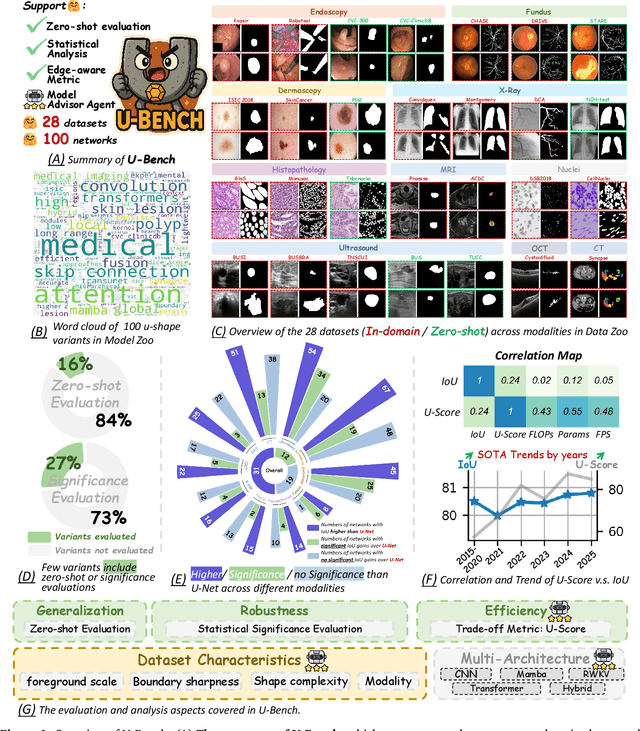


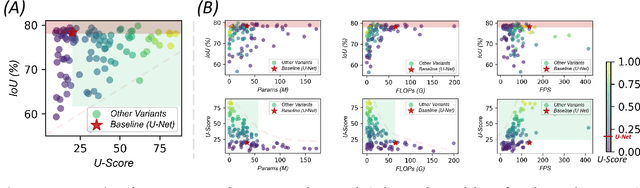
Abstract:Over the past decade, U-Net has been the dominant architecture in medical image segmentation, leading to the development of thousands of U-shaped variants. Despite its widespread adoption, there is still no comprehensive benchmark to systematically evaluate their performance and utility, largely because of insufficient statistical validation and limited consideration of efficiency and generalization across diverse datasets. To bridge this gap, we present U-Bench, the first large-scale, statistically rigorous benchmark that evaluates 100 U-Net variants across 28 datasets and 10 imaging modalities. Our contributions are threefold: (1) Comprehensive Evaluation: U-Bench evaluates models along three key dimensions: statistical robustness, zero-shot generalization, and computational efficiency. We introduce a novel metric, U-Score, which jointly captures the performance-efficiency trade-off, offering a deployment-oriented perspective on model progress. (2) Systematic Analysis and Model Selection Guidance: We summarize key findings from the large-scale evaluation and systematically analyze the impact of dataset characteristics and architectural paradigms on model performance. Based on these insights, we propose a model advisor agent to guide researchers in selecting the most suitable models for specific datasets and tasks. (3) Public Availability: We provide all code, models, protocols, and weights, enabling the community to reproduce our results and extend the benchmark with future methods. In summary, U-Bench not only exposes gaps in previous evaluations but also establishes a foundation for fair, reproducible, and practically relevant benchmarking in the next decade of U-Net-based segmentation models. The project can be accessed at: https://fenghetan9.github.io/ubench. Code is available at: https://github.com/FengheTan9/U-Bench.
A General Knowledge Injection Framework for ICD Coding
May 24, 2025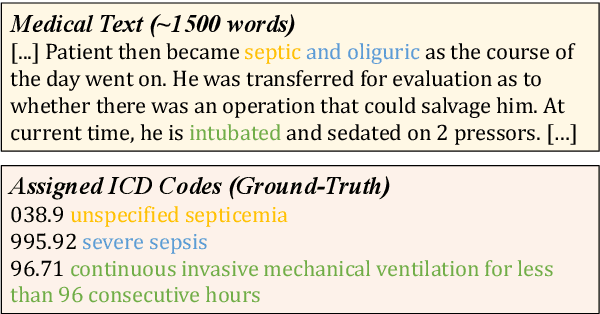
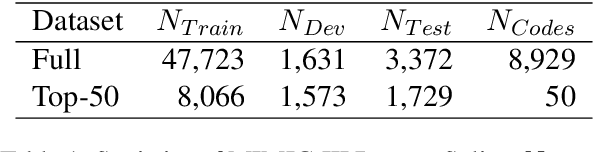
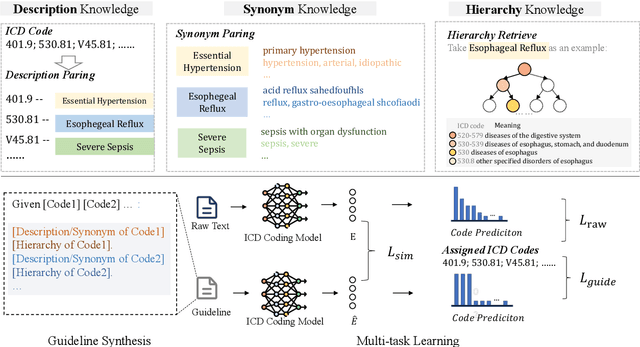
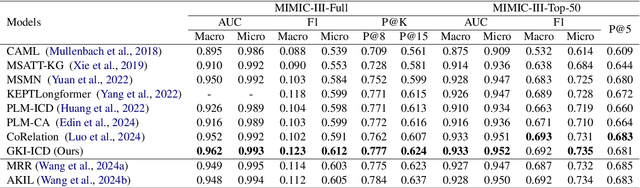
Abstract:ICD Coding aims to assign a wide range of medical codes to a medical text document, which is a popular and challenging task in the healthcare domain. To alleviate the problems of long-tail distribution and the lack of annotations of code-specific evidence, many previous works have proposed incorporating code knowledge to improve coding performance. However, existing methods often focus on a single type of knowledge and design specialized modules that are complex and incompatible with each other, thereby limiting their scalability and effectiveness. To address this issue, we propose GKI-ICD, a novel, general knowledge injection framework that integrates three key types of knowledge, namely ICD Description, ICD Synonym, and ICD Hierarchy, without specialized design of additional modules. The comprehensive utilization of the above knowledge, which exhibits both differences and complementarity, can effectively enhance the ICD coding performance. Extensive experiments on existing popular ICD coding benchmarks demonstrate the effectiveness of GKI-ICD, which achieves the state-of-the-art performance on most evaluation metrics. Code is available at https://github.com/xuzhang0112/GKI-ICD.
AA-CLIP: Enhancing Zero-shot Anomaly Detection via Anomaly-Aware CLIP
Mar 09, 2025Abstract:Anomaly detection (AD) identifies outliers for applications like defect and lesion detection. While CLIP shows promise for zero-shot AD tasks due to its strong generalization capabilities, its inherent Anomaly-Unawareness leads to limited discrimination between normal and abnormal features. To address this problem, we propose Anomaly-Aware CLIP (AA-CLIP), which enhances CLIP's anomaly discrimination ability in both text and visual spaces while preserving its generalization capability. AA-CLIP is achieved through a straightforward yet effective two-stage approach: it first creates anomaly-aware text anchors to differentiate normal and abnormal semantics clearly, then aligns patch-level visual features with these anchors for precise anomaly localization. This two-stage strategy, with the help of residual adapters, gradually adapts CLIP in a controlled manner, achieving effective AD while maintaining CLIP's class knowledge. Extensive experiments validate AA-CLIP as a resource-efficient solution for zero-shot AD tasks, achieving state-of-the-art results in industrial and medical applications. The code is available at https://github.com/Mwxinnn/AA-CLIP.
* 8 pages, 7 figures
Hi-End-MAE: Hierarchical encoder-driven masked autoencoders are stronger vision learners for medical image segmentation
Feb 12, 2025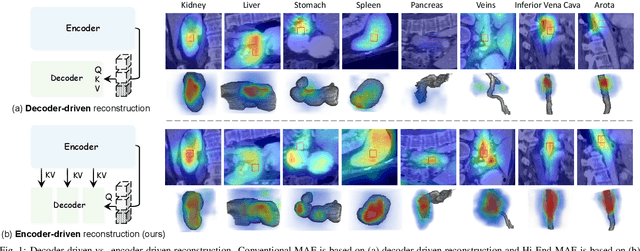
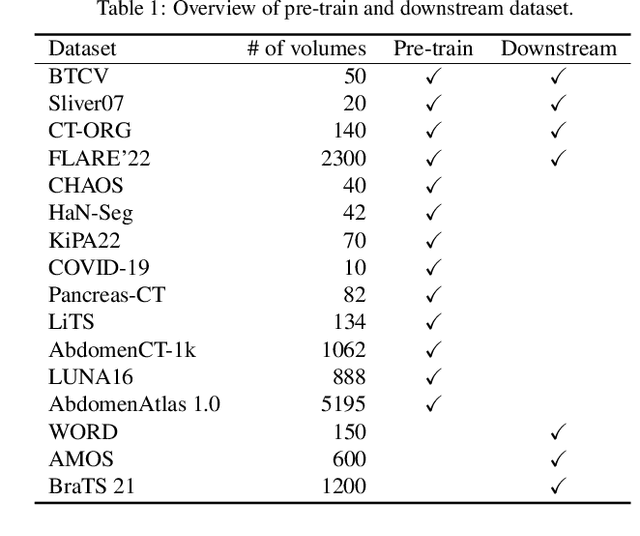
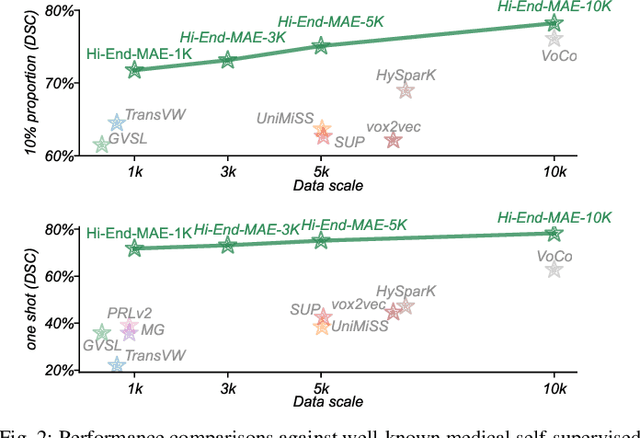
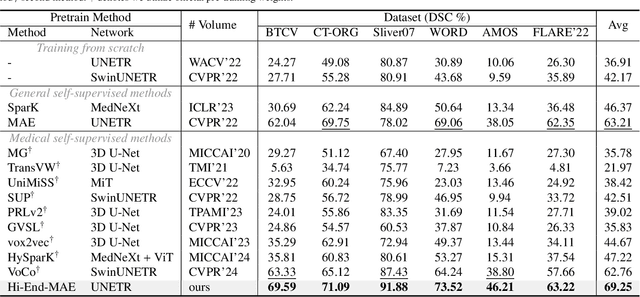
Abstract:Medical image segmentation remains a formidable challenge due to the label scarcity. Pre-training Vision Transformer (ViT) through masked image modeling (MIM) on large-scale unlabeled medical datasets presents a promising solution, providing both computational efficiency and model generalization for various downstream tasks. However, current ViT-based MIM pre-training frameworks predominantly emphasize local aggregation representations in output layers and fail to exploit the rich representations across different ViT layers that better capture fine-grained semantic information needed for more precise medical downstream tasks. To fill the above gap, we hereby present Hierarchical Encoder-driven MAE (Hi-End-MAE), a simple yet effective ViT-based pre-training solution, which centers on two key innovations: (1) Encoder-driven reconstruction, which encourages the encoder to learn more informative features to guide the reconstruction of masked patches; and (2) Hierarchical dense decoding, which implements a hierarchical decoding structure to capture rich representations across different layers. We pre-train Hi-End-MAE on a large-scale dataset of 10K CT scans and evaluated its performance across seven public medical image segmentation benchmarks. Extensive experiments demonstrate that Hi-End-MAE achieves superior transfer learning capabilities across various downstream tasks, revealing the potential of ViT in medical imaging applications. The code is available at: https://github.com/FengheTan9/Hi-End-MAE
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge