Qiong He
Single PW takes a shortcut to compound PW in US imaging
Dec 15, 2023Abstract:Reconstruction of ultrasound (US) images from radio-frequency data can be conceptualized as a linear inverse problem. Traditional deep learning approaches, which aim to improve the quality of US images by directly learning priors, often encounter challenges in generalization. Recently, diffusion-based generative models have received significant attention within the research community due to their robust performance in image reconstruction tasks. However, a limitation of these models is their inherent low speed in generating image samples from pure Gaussian noise progressively. In this study, we exploit the inherent similarity between the US images reconstructed from a single plane wave (PW) and PW compounding PWC). We hypothesize that a single PW can take a shortcut to reach the diffusion trajectory of PWC, removing the need to begin with Gaussian noise. By employing an advanced diffusion model, we demonstrate its effectiveness in US image reconstruction, achieving a substantial reduction in sampling steps. In-vivo experimental results indicate that our approach can reduce sampling steps by 60%, while preserving comparable performance metrics with the conventional diffusion model.
Fast Sampling generative model for Ultrasound image reconstruction
Dec 15, 2023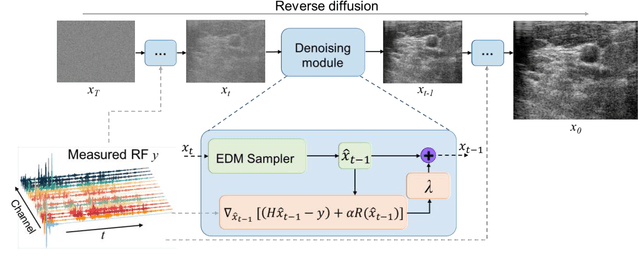

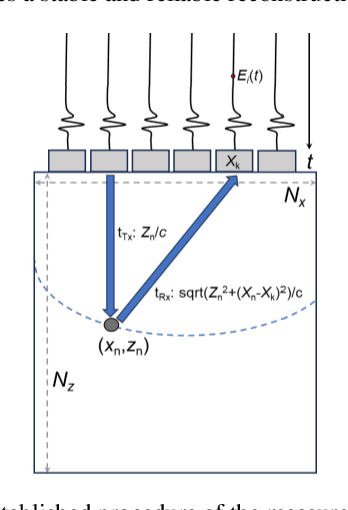
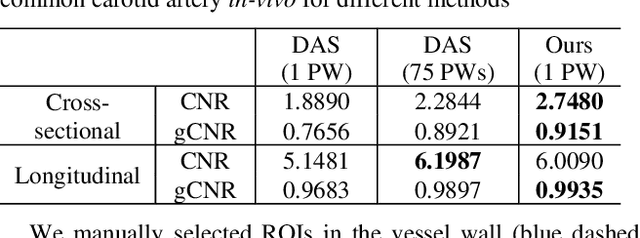
Abstract:Image reconstruction from radio-frequency data is pivotal in ultrafast plane wave ultrasound imaging. Unlike the conventional delay-and-sum (DAS) technique, which relies on somewhat imprecise assumptions, deep learning-based methods perform image reconstruction by training on paired data, leading to a notable enhancement in image quality. Nevertheless, these strategies often exhibit limited generalization capabilities. Recently, denoising diffusion models have become the preferred paradigm for image reconstruction tasks. However, their reliance on an iterative sampling procedure results in prolonged generation time. In this paper, we propose a novel sampling framework that concurrently enforces data consistency of ultrasound signals and data-driven priors. By leveraging the advanced diffusion model, the generation of high-quality images is substantially expedited. Experimental evaluations on an in-vivo dataset indicate that our approach with a single plane wave surpasses DAS with spatial coherent compounding of 75 plane waves.
A semi-automatic ultrasound image analysis system for the grading diagnosis of COVID-19 pneumonia
Nov 04, 2021


Abstract:This paper proposes a semi-automatic system based on quantitative characterization of the specific image patterns in lung ultrasound (LUS) images, in order to assess the lung conditions of patients with COVID-19 pneumonia, as well as to differentiate between the severe / and no-severe cases. Specifically, four parameters are extracted from each LUS image, namely the thickness (TPL) and roughness (RPL) of the pleural line, and the accumulated with (AWBL) and acoustic coefficient (ACBL) of B lines. 27 patients are enrolled in this study, which are grouped into 13 moderate patients, 7 severe patients and 7 critical patients. Furthermore, the severe and critical patients are regarded as the severe cases, and the moderate patients are regarded as the non-severe cases. Biomarkers among different groups are compared. Each single biomarker and a classifier with all the biomarkers as input are utilized for the binary diagnosis of severe case and non-severe case, respectively. The classifier achieves the best classification performance among all the compared methods (area under the receiver operating characteristics curve = 0.93, sensitivity = 0.93, specificity = 0.85). The proposed image analysis system could be potentially applied to the grading and prognosis evaluation of patients with COVID-19 pneumonia.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge