Ruiyang Li
SkinFlow: Efficient Information Transmission for Open Dermatological Diagnosis via Dynamic Visual Encoding and Staged RL
Jan 14, 2026Abstract:General-purpose Large Vision-Language Models (LVLMs), despite their massive scale, often falter in dermatology due to "diffuse attention" - the inability to disentangle subtle pathological lesions from background noise. In this paper, we challenge the assumption that parameter scaling is the only path to medical precision. We introduce SkinFlow, a framework that treats diagnosis as an optimization of visual information transmission efficiency. Our approach utilizes a Virtual-Width Dynamic Vision Encoder (DVE) to "unfold" complex pathological manifolds without physical parameter expansion, coupled with a two-stage Reinforcement Learning strategy. This strategy sequentially aligns explicit medical descriptions (Stage I) and reconstructs implicit diagnostic textures (Stage II) within a constrained semantic space. Furthermore, we propose a clinically grounded evaluation protocol that prioritizes diagnostic safety and hierarchical relevance over rigid label matching. Empirical results are compelling: our 7B model establishes a new state-of-the-art on the Fitzpatrick17k benchmark, achieving a +12.06% gain in Top-1 accuracy and a +28.57% boost in Top-6 accuracy over the massive general-purpose models (e.g., Qwen3VL-235B and GPT-5.2). These findings demonstrate that optimizing geometric capacity and information flow yields superior diagnostic reasoning compared to raw parameter scaling.
ClipGS: Clippable Gaussian Splatting for Interactive Cinematic Visualization of Volumetric Medical Data
Jul 09, 2025Abstract:The visualization of volumetric medical data is crucial for enhancing diagnostic accuracy and improving surgical planning and education. Cinematic rendering techniques significantly enrich this process by providing high-quality visualizations that convey intricate anatomical details, thereby facilitating better understanding and decision-making in medical contexts. However, the high computing cost and low rendering speed limit the requirement of interactive visualization in practical applications. In this paper, we introduce ClipGS, an innovative Gaussian splatting framework with the clipping plane supported, for interactive cinematic visualization of volumetric medical data. To address the challenges posed by dynamic interactions, we propose a learnable truncation scheme that automatically adjusts the visibility of Gaussian primitives in response to the clipping plane. Besides, we also design an adaptive adjustment model to dynamically adjust the deformation of Gaussians and refine the rendering performance. We validate our method on five volumetric medical data (including CT and anatomical slice data), and reach an average 36.635 PSNR rendering quality with 156 FPS and 16.1 MB model size, outperforming state-of-the-art methods in rendering quality and efficiency.
Efficient Medical VIE via Reinforcement Learning
Jun 16, 2025Abstract:Visual Information Extraction (VIE) converts unstructured document images into structured formats like JSON, critical for medical applications such as report analysis and online consultations. Traditional methods rely on OCR and language models, while end-to-end multimodal models offer direct JSON generation. However, domain-specific schemas and high annotation costs limit their effectiveness in medical VIE. We base our approach on the Reinforcement Learning with Verifiable Rewards (RLVR) framework to address these challenges using only 100 annotated samples. Our approach ensures dataset diversity, a balanced precision-recall reward mechanism to reduce hallucinations and improve field coverage, and innovative sampling strategies to enhance reasoning capabilities. Fine-tuning Qwen2.5-VL-7B with our RLVR method, we achieve state-of-the-art performance on medical VIE tasks, significantly improving F1, precision, and recall. While our models excel on tasks similar to medical datasets, performance drops on dissimilar tasks, highlighting the need for domain-specific optimization. Case studies further demonstrate the value of reasoning during training and inference for VIE.
MS2Mesh-XR: Multi-modal Sketch-to-Mesh Generation in XR Environments
Dec 12, 2024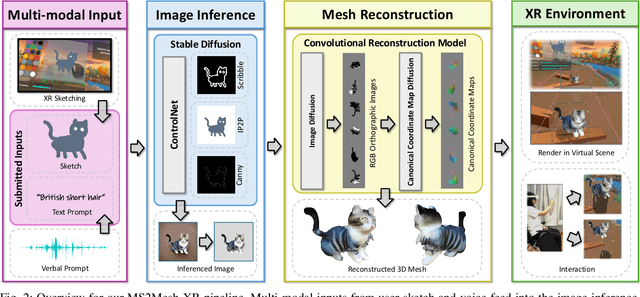
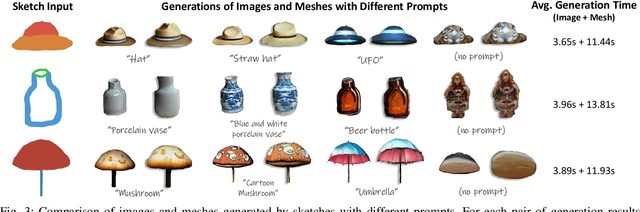
Abstract:We present MS2Mesh-XR, a novel multi-modal sketch-to-mesh generation pipeline that enables users to create realistic 3D objects in extended reality (XR) environments using hand-drawn sketches assisted by voice inputs. In specific, users can intuitively sketch objects using natural hand movements in mid-air within a virtual environment. By integrating voice inputs, we devise ControlNet to infer realistic images based on the drawn sketches and interpreted text prompts. Users can then review and select their preferred image, which is subsequently reconstructed into a detailed 3D mesh using the Convolutional Reconstruction Model. In particular, our proposed pipeline can generate a high-quality 3D mesh in less than 20 seconds, allowing for immersive visualization and manipulation in run-time XR scenes. We demonstrate the practicability of our pipeline through two use cases in XR settings. By leveraging natural user inputs and cutting-edge generative AI capabilities, our approach can significantly facilitate XR-based creative production and enhance user experiences. Our code and demo will be available at: https://yueqiu0911.github.io/MS2Mesh-XR/
Full Information Linked ICA: addressing missing data problem in multimodal fusion
Jun 27, 2024



Abstract:Recent advances in multimodal imaging acquisition techniques have allowed us to measure different aspects of brain structure and function. Multimodal fusion, such as linked independent component analysis (LICA), is popularly used to integrate complementary information. However, it has suffered from missing data, commonly occurring in neuroimaging data. Therefore, in this paper, we propose a Full Information LICA algorithm (FI-LICA) to handle the missing data problem during multimodal fusion under the LICA framework. Built upon complete cases, our method employs the principle of full information and utilizes all available information to recover the missing latent information. Our simulation experiments showed the ideal performance of FI-LICA compared to current practices. Further, we applied FI-LICA to multimodal data from the Alzheimer's Disease Neuroimaging Initiative (ADNI) study, showcasing better performance in classifying current diagnosis and in predicting the AD transition of participants with mild cognitive impairment (MCI), thereby highlighting the practical utility of our proposed method.
HGT: A Hierarchical GCN-Based Transformer for Multimodal Periprosthetic Joint Infection Diagnosis Using CT Images and Text
May 29, 2023



Abstract:Prosthetic Joint Infection (PJI) is a prevalent and severe complication characterized by high diagnostic challenges. Currently, a unified diagnostic standard incorporating both computed tomography (CT) images and numerical text data for PJI remains unestablished, owing to the substantial noise in CT images and the disparity in data volume between CT images and text data. This study introduces a diagnostic method, HGT, based on deep learning and multimodal techniques. It effectively merges features from CT scan images and patients' numerical text data via a Unidirectional Selective Attention (USA) mechanism and a graph convolutional network (GCN)-based feature fusion network. We evaluated the proposed method on a custom-built multimodal PJI dataset, assessing its performance through ablation experiments and interpretability evaluations. Our method achieved an accuracy (ACC) of 91.4\% and an area under the curve (AUC) of 95.9\%, outperforming recent multimodal approaches by 2.9\% in ACC and 2.2\% in AUC, with a parameter count of only 68M. Notably, the interpretability results highlighted our model's strong focus and localization capabilities at lesion sites. This proposed method could provide clinicians with additional diagnostic tools to enhance accuracy and efficiency in clinical practice.
Cascaded Feature Warping Network for Unsupervised Medical Image Registration
Mar 15, 2021
Abstract:Deformable image registration is widely utilized in medical image analysis, but most proposed methods fail in the situation of complex deformations. In this paper, we pre-sent a cascaded feature warping network to perform the coarse-to-fine registration. To achieve this, a shared-weights encoder network is adopted to generate the feature pyramids for the unaligned images. The feature warping registration module is then used to estimate the deformation field at each level. The coarse-to-fine manner is implemented by cascading the module from the bottom level to the top level. Furthermore, the multi-scale loss is also introduced to boost the registration performance. We employ two public benchmark datasets and conduct various experiments to evaluate our method. The results show that our method outperforms the state-of-the-art methods, which also demonstrates that the cascaded feature warping network can perform the coarse-to-fine registration effectively and efficiently.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge