Joyce C. Ho
FusionDP: Foundation Model-Assisted Differentially Private Learning for Partially Sensitive Features
Nov 05, 2025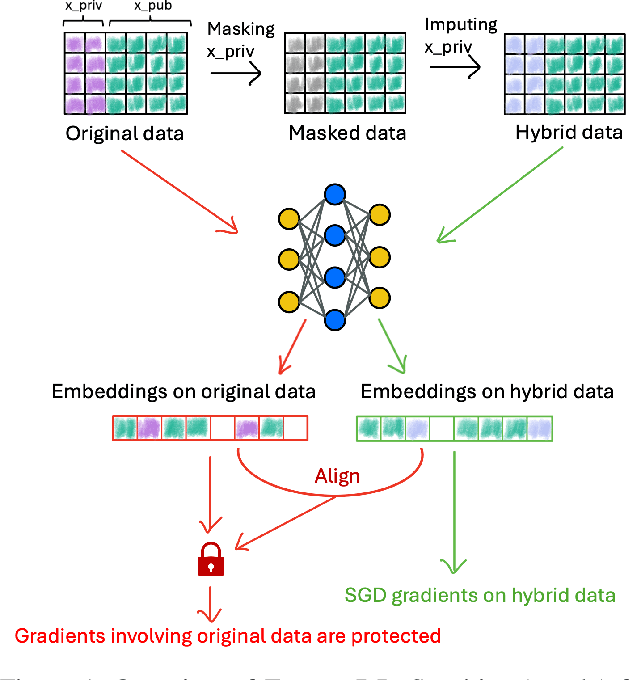
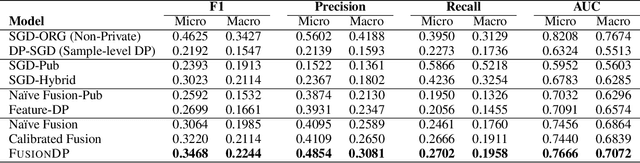
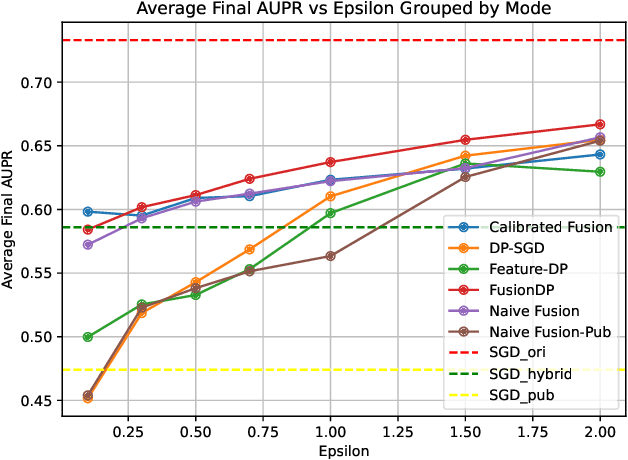
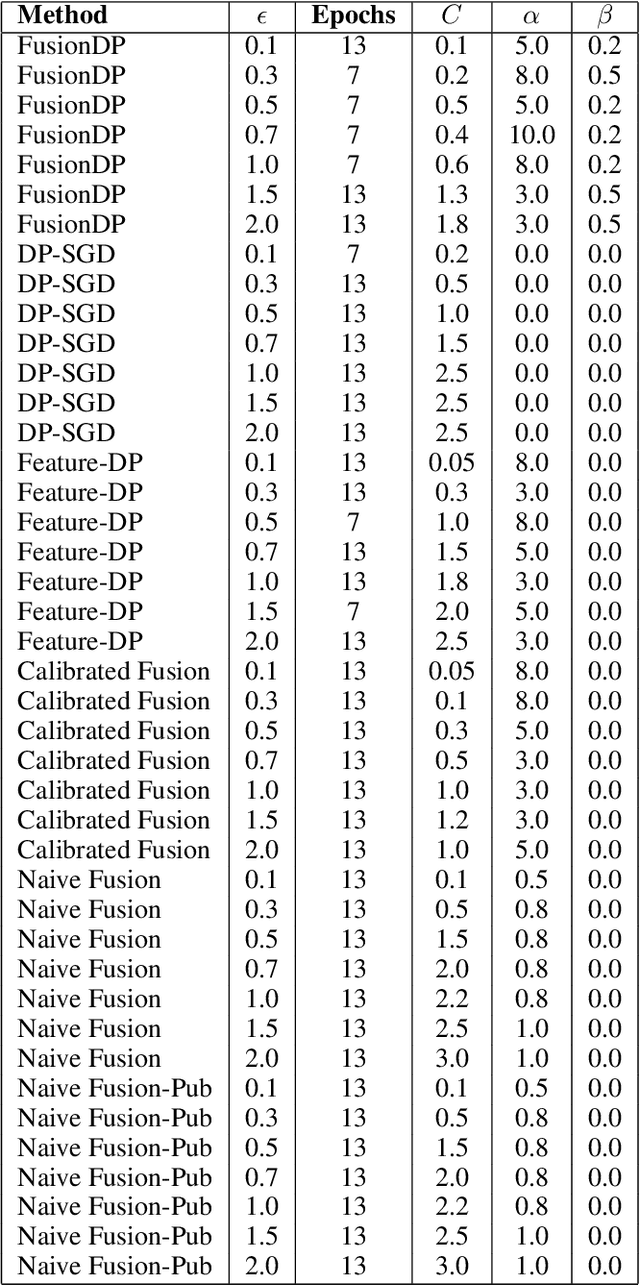
Abstract:Ensuring the privacy of sensitive training data is crucial in privacy-preserving machine learning. However, in practical scenarios, privacy protection may be required for only a subset of features. For instance, in ICU data, demographic attributes like age and gender pose higher privacy risks due to their re-identification potential, whereas raw lab results are generally less sensitive. Traditional DP-SGD enforces privacy protection on all features in one sample, leading to excessive noise injection and significant utility degradation. We propose FusionDP, a two-step framework that enhances model utility under feature-level differential privacy. First, FusionDP leverages large foundation models to impute sensitive features given non-sensitive features, treating them as external priors that provide high-quality estimates of sensitive attributes without accessing the true values during model training. Second, we introduce a modified DP-SGD algorithm that trains models on both original and imputed features while formally preserving the privacy of the original sensitive features. We evaluate FusionDP on two modalities: a sepsis prediction task on tabular data from PhysioNet and a clinical note classification task from MIMIC-III. By comparing against privacy-preserving baselines, our results show that FusionDP significantly improves model performance while maintaining rigorous feature-level privacy, demonstrating the potential of foundation model-driven imputation to enhance the privacy-utility trade-off for various modalities.
Collab-RAG: Boosting Retrieval-Augmented Generation for Complex Question Answering via White-Box and Black-Box LLM Collaboration
Apr 07, 2025Abstract:Retrieval-Augmented Generation (RAG) systems often struggle to handle multi-hop question-answering tasks accurately due to irrelevant context retrieval and limited complex reasoning capabilities. We introduce Collab-RAG, a collaborative training framework that leverages mutual enhancement between a white-box small language model (SLM) and a blackbox large language model (LLM) for RAG. Specifically, the SLM decomposes complex queries into simpler sub-questions, thus enhancing the accuracy of the retrieval and facilitating more effective reasoning by the black-box LLM. Concurrently, the black-box LLM provides feedback signals to improve the SLM's decomposition capability. We observe that Collab-RAG relies solely on supervision from an affordable black-box LLM without additional distillation from frontier LLMs, yet demonstrates strong generalization across multiple black-box LLMs. Experimental evaluations across five multi-hop QA datasets demonstrate that Collab-RAG substantially outperforms existing black-box-only and SLM fine-tuning baselines by 1.8%-14.2% on average. In particular, our fine-tuned 3B SLM surpasses a frozen 32B LLM in question decomposition, highlighting the efficiency of Collab-RAG in improving reasoning and retrieval for complex questions. The code of Collab-RAG is available on https://github.com/ritaranx/Collab-RAG/.
Is thermography a viable solution for detecting pressure injuries in dark skin patients?
Nov 15, 2024Abstract:Pressure injury (PI) detection is challenging, especially in dark skin tones, due to the unreliability of visual inspection. Thermography has been suggested as a viable alternative as temperature differences in the skin can indicate impending tissue damage. Although deep learning models have demonstrated considerable promise toward reliably detecting PI, the existing work fails to evaluate the performance on darker skin tones and varying data collection protocols. In this paper, we introduce a new thermal and optical imaging dataset of 35 participants focused on darker skin tones where temperature differences are induced through cooling and cupping protocols. We vary the image collection process to include different cameras, lighting, patient pose, and camera distance. We compare the performance of a small convolutional neural network (CNN) trained on either the thermal or the optical images on all skin tones. Our preliminary results suggest that thermography-based CNN is robust to data collection protocols for all skin tones.
SimRAG: Self-Improving Retrieval-Augmented Generation for Adapting Large Language Models to Specialized Domains
Oct 23, 2024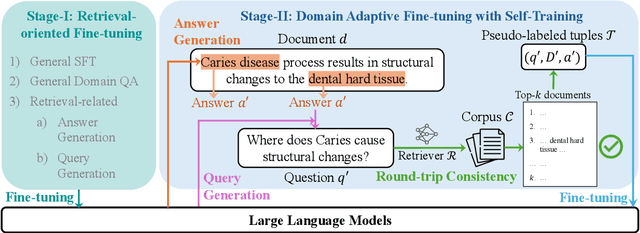
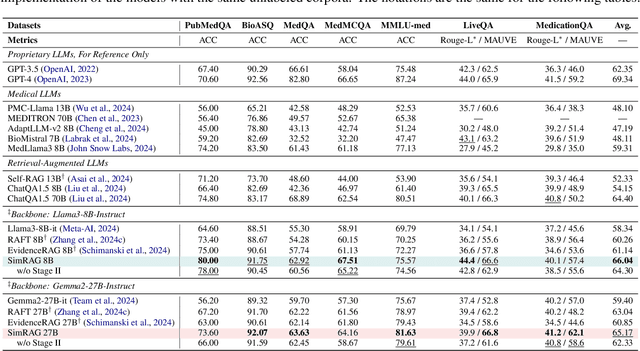
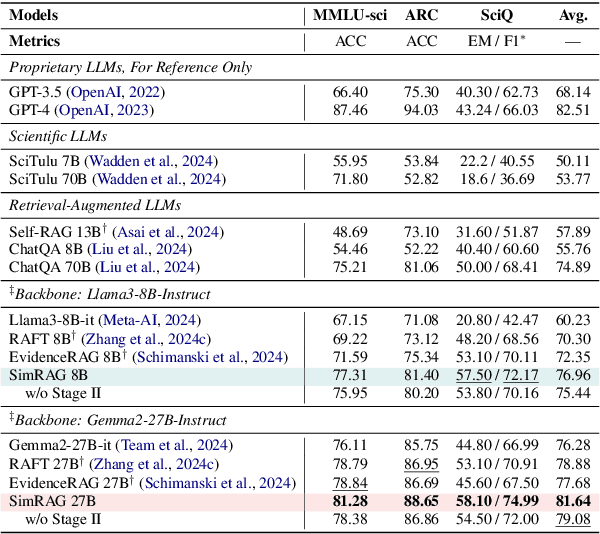
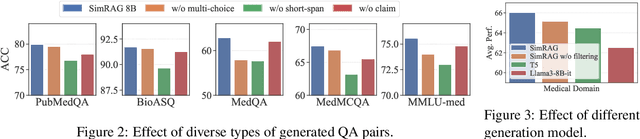
Abstract:Retrieval-augmented generation (RAG) enhances the question-answering (QA) abilities of large language models (LLMs) by integrating external knowledge. However, adapting general-purpose RAG systems to specialized fields such as science and medicine poses unique challenges due to distribution shifts and limited access to domain-specific data. To tackle this, we propose SimRAG, a self-training approach that equips the LLM with joint capabilities of question answering and question generation for domain adaptation. Our method first fine-tunes the LLM on instruction-following, question-answering, and search-related data. Then, it prompts the same LLM to generate diverse domain-relevant questions from unlabeled corpora, with an additional filtering strategy to retain high-quality synthetic examples. By leveraging these synthetic examples, the LLM can improve their performance on domain-specific RAG tasks. Experiments on 11 datasets, spanning two backbone sizes and three domains, demonstrate that SimRAG outperforms baselines by 1.2\%--8.6\%.
Large Language Models for Integrating Social Determinant of Health Data: A Case Study on Heart Failure 30-Day Readmission Prediction
Jul 12, 2024



Abstract:Social determinants of health (SDOH) $-$ the myriad of circumstances in which people live, grow, and age $-$ play an important role in health outcomes. However, existing outcome prediction models often only use proxies of SDOH as features. Recent open data initiatives present an opportunity to construct a more comprehensive view of SDOH, but manually integrating the most relevant data for individual patients becomes increasingly challenging as the volume and diversity of public SDOH data grows. Large language models (LLMs) have shown promise at automatically annotating structured data. Here, we conduct an end-to-end case study evaluating the feasibility of using LLMs to integrate SDOH data, and the utility of these SDOH features for clinical prediction. We first manually label 700+ variables from two publicly-accessible SDOH data sources to one of five semantic SDOH categories. Then, we benchmark performance of 9 open-source LLMs on this classification task. Finally, we train ML models to predict 30-day hospital readmission among 39k heart failure (HF) patients, and we compare the prediction performance of the categorized SDOH variables with standard clinical variables. Additionally, we investigate the impact of few-shot LLM prompting on LLM annotation performance, and perform a metadata ablation study on prompts to evaluate which information helps LLMs accurately annotate these variables. We find that some open-source LLMs can effectively, accurately annotate SDOH variables with zero-shot prompting without the need for fine-tuning. Crucially, when combined with standard clinical features, the LLM-annotated Neighborhood and Built Environment subset of the SDOH variables shows the best performance predicting 30-day readmission of HF patients.
TACCO: Task-guided Co-clustering of Clinical Concepts and Patient Visits for Disease Subtyping based on EHR Data
Jun 14, 2024



Abstract:The growing availability of well-organized Electronic Health Records (EHR) data has enabled the development of various machine learning models towards disease risk prediction. However, existing risk prediction methods overlook the heterogeneity of complex diseases, failing to model the potential disease subtypes regarding their corresponding patient visits and clinical concept subgroups. In this work, we introduce TACCO, a novel framework that jointly discovers clusters of clinical concepts and patient visits based on a hypergraph modeling of EHR data. Specifically, we develop a novel self-supervised co-clustering framework that can be guided by the risk prediction task of specific diseases. Furthermore, we enhance the hypergraph model of EHR data with textual embeddings and enforce the alignment between the clusters of clinical concepts and patient visits through a contrastive objective. Comprehensive experiments conducted on the public MIMIC-III dataset and Emory internal CRADLE dataset over the downstream clinical tasks of phenotype classification and cardiovascular risk prediction demonstrate an average 31.25% performance improvement compared to traditional ML baselines and a 5.26% improvement on top of the vanilla hypergraph model without our co-clustering mechanism. In-depth model analysis, clustering results analysis, and clinical case studies further validate the improved utilities and insightful interpretations delivered by TACCO. Code is available at https://github.com/PericlesHat/TACCO.
BMRetriever: Tuning Large Language Models as Better Biomedical Text Retrievers
Apr 29, 2024



Abstract:Developing effective biomedical retrieval models is important for excelling at knowledge-intensive biomedical tasks but still challenging due to the deficiency of sufficient publicly annotated biomedical data and computational resources. We present BMRetriever, a series of dense retrievers for enhancing biomedical retrieval via unsupervised pre-training on large biomedical corpora, followed by instruction fine-tuning on a combination of labeled datasets and synthetic pairs. Experiments on 5 biomedical tasks across 11 datasets verify BMRetriever's efficacy on various biomedical applications. BMRetriever also exhibits strong parameter efficiency, with the 410M variant outperforming baselines up to 11.7 times larger, and the 2B variant matching the performance of models with over 5B parameters. The training data and model checkpoints are released at \url{https://huggingface.co/BMRetriever} to ensure transparency, reproducibility, and application to new domains.
LLMs-based Few-Shot Disease Predictions using EHR: A Novel Approach Combining Predictive Agent Reasoning and Critical Agent Instruction
Mar 19, 2024Abstract:Electronic health records (EHRs) contain valuable patient data for health-related prediction tasks, such as disease prediction. Traditional approaches rely on supervised learning methods that require large labeled datasets, which can be expensive and challenging to obtain. In this study, we investigate the feasibility of applying Large Language Models (LLMs) to convert structured patient visit data (e.g., diagnoses, labs, prescriptions) into natural language narratives. We evaluate the zero-shot and few-shot performance of LLMs using various EHR-prediction-oriented prompting strategies. Furthermore, we propose a novel approach that utilizes LLM agents with different roles: a predictor agent that makes predictions and generates reasoning processes and a critic agent that analyzes incorrect predictions and provides guidance for improving the reasoning of the predictor agent. Our results demonstrate that with the proposed approach, LLMs can achieve decent few-shot performance compared to traditional supervised learning methods in EHR-based disease predictions, suggesting its potential for health-oriented applications.
RAM-EHR: Retrieval Augmentation Meets Clinical Predictions on Electronic Health Records
Feb 25, 2024Abstract:We present RAM-EHR, a Retrieval AugMentation pipeline to improve clinical predictions on Electronic Health Records (EHRs). RAM-EHR first collects multiple knowledge sources, converts them into text format, and uses dense retrieval to obtain information related to medical concepts. This strategy addresses the difficulties associated with complex names for the concepts. RAM-EHR then augments the local EHR predictive model co-trained with consistency regularization to capture complementary information from patient visits and summarized knowledge. Experiments on two EHR datasets show the efficacy of RAM-EHR over previous knowledge-enhanced baselines (3.4% gain in AUROC and 7.2% gain in AUPR), emphasizing the effectiveness of the summarized knowledge from RAM-EHR for clinical prediction tasks. The code will be published at \url{https://github.com/ritaranx/RAM-EHR}.
A Deep Reinforcement Learning Approach for Interactive Search with Sentence-level Feedback
Oct 03, 2023Abstract:Interactive search can provide a better experience by incorporating interaction feedback from the users. This can significantly improve search accuracy as it helps avoid irrelevant information and captures the users' search intents. Existing state-of-the-art (SOTA) systems use reinforcement learning (RL) models to incorporate the interactions but focus on item-level feedback, ignoring the fine-grained information found in sentence-level feedback. Yet such feedback requires extensive RL action space exploration and large amounts of annotated data. This work addresses these challenges by proposing a new deep Q-learning (DQ) approach, DQrank. DQrank adapts BERT-based models, the SOTA in natural language processing, to select crucial sentences based on users' engagement and rank the items to obtain more satisfactory responses. We also propose two mechanisms to better explore optimal actions. DQrank further utilizes the experience replay mechanism in DQ to store the feedback sentences to obtain a better initial ranking performance. We validate the effectiveness of DQrank on three search datasets. The results show that DQRank performs at least 12% better than the previous SOTA RL approaches. We also conduct detailed ablation studies. The ablation results demonstrate that each model component can efficiently extract and accumulate long-term engagement effects from the users' sentence-level feedback. This structure offers new technologies with promised performance to construct a search system with sentence-level interaction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge