May D. Wang
LLM-as-RNN: A Recurrent Language Model for Memory Updates and Sequence Prediction
Jan 19, 2026Abstract:Large language models are strong sequence predictors, yet standard inference relies on immutable context histories. After making an error at generation step t, the model lacks an updatable memory mechanism that improves predictions for step t+1. We propose LLM-as-RNN, an inference-only framework that turns a frozen LLM into a recurrent predictor by representing its hidden state as natural-language memory. This state, implemented as a structured system-prompt summary, is updated at each timestep via feedback-driven text rewrites, enabling learning without parameter updates. Under a fixed token budget, LLM-as-RNN corrects errors and retains task-relevant patterns, effectively performing online learning through language. We evaluate the method on three sequential benchmarks in healthcare, meteorology, and finance across Llama, Gemma, and GPT model families. LLM-as-RNN significantly outperforms zero-shot, full-history, and MemPrompt baselines, improving predictive accuracy by 6.5% on average, while producing interpretable, human-readable learning traces absent in standard context accumulation.
MedAgentGym: Training LLM Agents for Code-Based Medical Reasoning at Scale
Jun 04, 2025



Abstract:We introduce MedAgentGYM, the first publicly available training environment designed to enhance coding-based medical reasoning capabilities in large language model (LLM) agents. MedAgentGYM comprises 72,413 task instances across 129 categories derived from authentic real-world biomedical scenarios. Tasks are encapsulated within executable coding environments, each featuring detailed task descriptions, interactive feedback mechanisms, verifiable ground-truth annotations, and scalable training trajectory generation. Extensive benchmarking of over 30 LLMs reveals a notable performance disparity between commercial API-based models and open-source counterparts. Leveraging MedAgentGYM, Med-Copilot-7B achieves substantial performance gains through supervised fine-tuning (+36.44%) and continued reinforcement learning (+42.47%), emerging as an affordable and privacy-preserving alternative competitive with gpt-4o. By offering both a comprehensive benchmark and accessible, expandable training resources within unified execution environments, MedAgentGYM delivers an integrated platform to develop LLM-based coding assistants for advanced biomedical research and practice.
Novel Extraction of Discriminative Fine-Grained Feature to Improve Retinal Vessel Segmentation
May 06, 2025Abstract:Retinal vessel segmentation is a vital early detection method for several severe ocular diseases. Despite significant progress in retinal vessel segmentation with the advancement of Neural Networks, there are still challenges to overcome. Specifically, retinal vessel segmentation aims to predict the class label for every pixel within a fundus image, with a primary focus on intra-image discrimination, making it vital for models to extract more discriminative features. Nevertheless, existing methods primarily focus on minimizing the difference between the output from the decoder and the label, but ignore fully using feature-level fine-grained representations from the encoder. To address these issues, we propose a novel Attention U-shaped Kolmogorov-Arnold Network named AttUKAN along with a novel Label-guided Pixel-wise Contrastive Loss for retinal vessel segmentation. Specifically, we implement Attention Gates into Kolmogorov-Arnold Networks to enhance model sensitivity by suppressing irrelevant feature activations and model interpretability by non-linear modeling of KAN blocks. Additionally, we also design a novel Label-guided Pixel-wise Contrastive Loss to supervise our proposed AttUKAN to extract more discriminative features by distinguishing between foreground vessel-pixel pairs and background pairs. Experiments are conducted across four public datasets including DRIVE, STARE, CHASE_DB1, HRF and our private dataset. AttUKAN achieves F1 scores of 82.50%, 81.14%, 81.34%, 80.21% and 80.09%, along with MIoU scores of 70.24%, 68.64%, 68.59%, 67.21% and 66.94% in the above datasets, which are the highest compared to 11 networks for retinal vessel segmentation. Quantitative and qualitative results show that our AttUKAN achieves state-of-the-art performance and outperforms existing retinal vessel segmentation methods. Our code will be available at https://github.com/stevezs315/AttUKAN.
Advancing Problem-Based Learning in Biomedical Engineering in the Era of Generative AI
Mar 20, 2025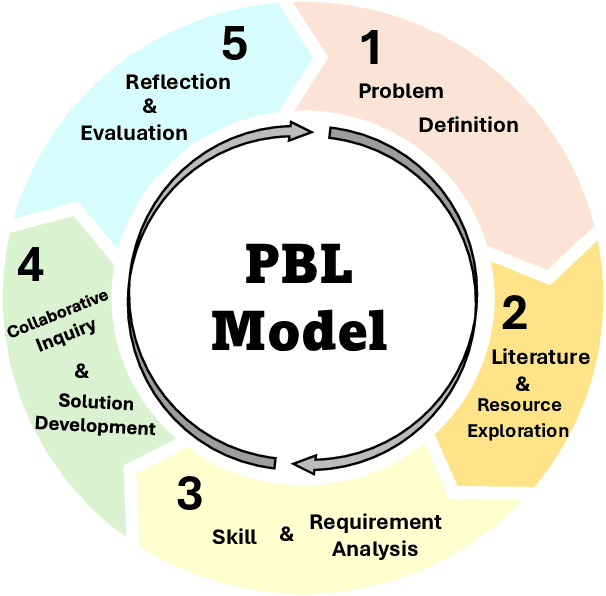
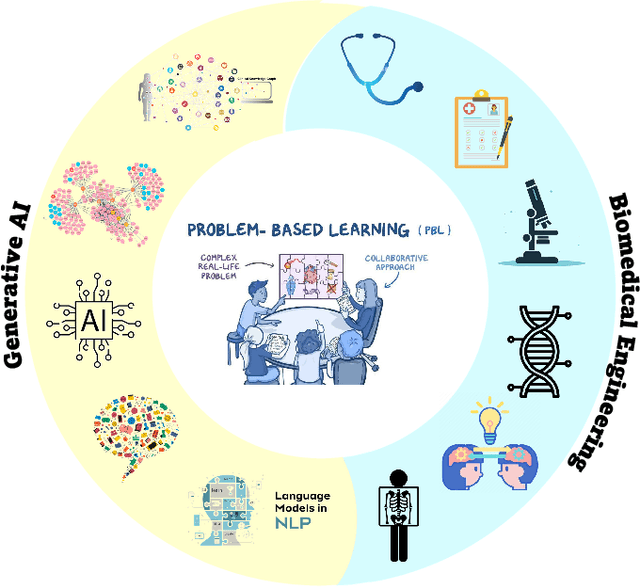

Abstract:Problem-Based Learning (PBL) has significantly impacted biomedical engineering (BME) education since its introduction in the early 2000s, effectively enhancing critical thinking and real-world knowledge application among students. With biomedical engineering rapidly converging with artificial intelligence (AI), integrating effective AI education into established curricula has become challenging yet increasingly necessary. Recent advancements, including AI's recognition by the 2024 Nobel Prize, have highlighted the importance of training students comprehensively in biomedical AI. However, effective biomedical AI education faces substantial obstacles, such as diverse student backgrounds, limited personalized mentoring, constrained computational resources, and difficulties in safely scaling hands-on practical experiments due to privacy and ethical concerns associated with biomedical data. To overcome these issues, we conducted a three-year (2021-2023) case study implementing an advanced PBL framework tailored specifically for biomedical AI education, involving 92 undergraduate and 156 graduate students from the joint Biomedical Engineering program of Georgia Institute of Technology and Emory University. Our approach emphasizes collaborative, interdisciplinary problem-solving through authentic biomedical AI challenges. The implementation led to measurable improvements in learning outcomes, evidenced by high research productivity (16 student-authored publications), consistently positive peer evaluations, and successful development of innovative computational methods addressing real biomedical challenges. Additionally, we examined the role of generative AI both as a teaching subject and an educational support tool within the PBL framework. Our study presents a practical and scalable roadmap for biomedical engineering departments aiming to integrate robust AI education into their curricula.
MedAdapter: Efficient Test-Time Adaptation of Large Language Models towards Medical Reasoning
May 05, 2024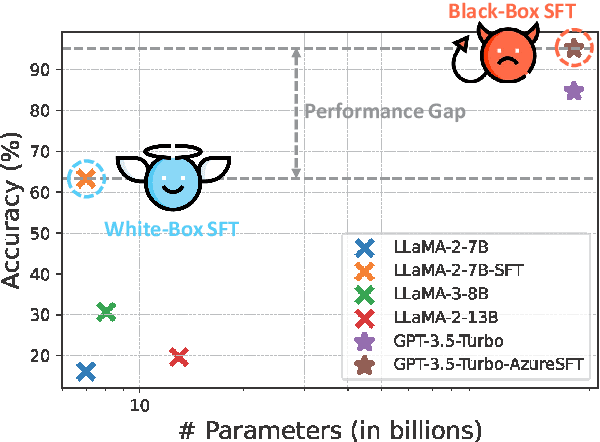
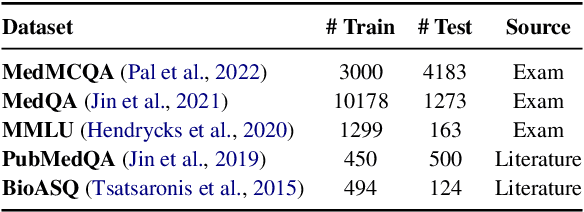
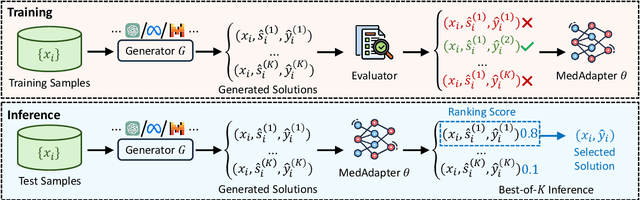
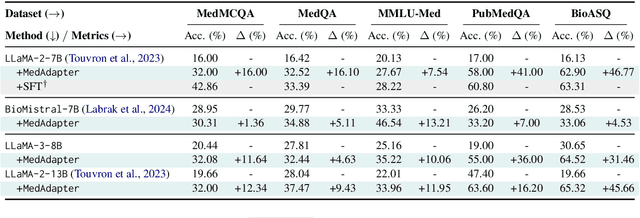
Abstract:Despite their improved capabilities in generation and reasoning, adapting large language models (LLMs) to the biomedical domain remains challenging due to their immense size and corporate privacy. In this work, we propose MedAdapter, a unified post-hoc adapter for test-time adaptation of LLMs towards biomedical applications. Instead of fine-tuning the entire LLM, MedAdapter effectively adapts the original model by fine-tuning only a small BERT-sized adapter to rank candidate solutions generated by LLMs. Experiments demonstrate that MedAdapter effectively adapts both white-box and black-box LLMs in biomedical reasoning, achieving average performance improvements of 25.48% and 11.31%, respectively, without requiring extensive computational resources or sharing data with third parties. MedAdapter also yields superior performance when combined with train-time adaptation, highlighting a flexible and complementary solution to existing adaptation methods. Faced with the challenges of balancing model performance, computational resources, and data privacy, MedAdapter provides an efficient, privacy-preserving, cost-effective, and transparent solution for adapting LLMs to the biomedical domain.
BMRetriever: Tuning Large Language Models as Better Biomedical Text Retrievers
Apr 29, 2024



Abstract:Developing effective biomedical retrieval models is important for excelling at knowledge-intensive biomedical tasks but still challenging due to the deficiency of sufficient publicly annotated biomedical data and computational resources. We present BMRetriever, a series of dense retrievers for enhancing biomedical retrieval via unsupervised pre-training on large biomedical corpora, followed by instruction fine-tuning on a combination of labeled datasets and synthetic pairs. Experiments on 5 biomedical tasks across 11 datasets verify BMRetriever's efficacy on various biomedical applications. BMRetriever also exhibits strong parameter efficiency, with the 410M variant outperforming baselines up to 11.7 times larger, and the 2B variant matching the performance of models with over 5B parameters. The training data and model checkpoints are released at \url{https://huggingface.co/BMRetriever} to ensure transparency, reproducibility, and application to new domains.
RAM-EHR: Retrieval Augmentation Meets Clinical Predictions on Electronic Health Records
Feb 25, 2024Abstract:We present RAM-EHR, a Retrieval AugMentation pipeline to improve clinical predictions on Electronic Health Records (EHRs). RAM-EHR first collects multiple knowledge sources, converts them into text format, and uses dense retrieval to obtain information related to medical concepts. This strategy addresses the difficulties associated with complex names for the concepts. RAM-EHR then augments the local EHR predictive model co-trained with consistency regularization to capture complementary information from patient visits and summarized knowledge. Experiments on two EHR datasets show the efficacy of RAM-EHR over previous knowledge-enhanced baselines (3.4% gain in AUROC and 7.2% gain in AUPR), emphasizing the effectiveness of the summarized knowledge from RAM-EHR for clinical prediction tasks. The code will be published at \url{https://github.com/ritaranx/RAM-EHR}.
EHRAgent: Code Empowers Large Language Models for Complex Tabular Reasoning on Electronic Health Records
Jan 13, 2024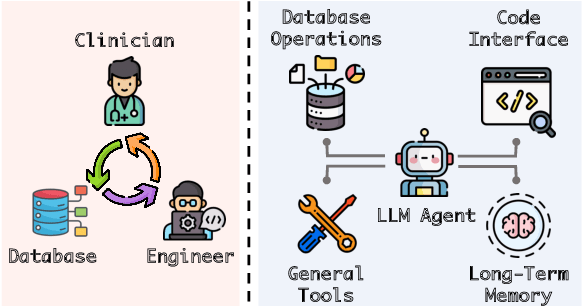
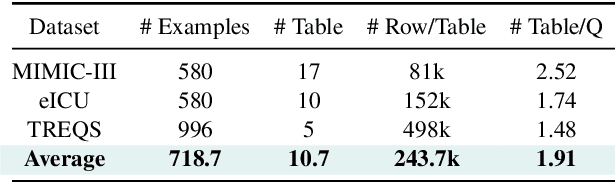
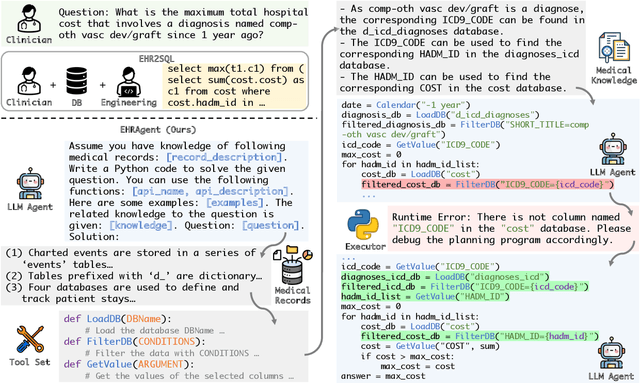
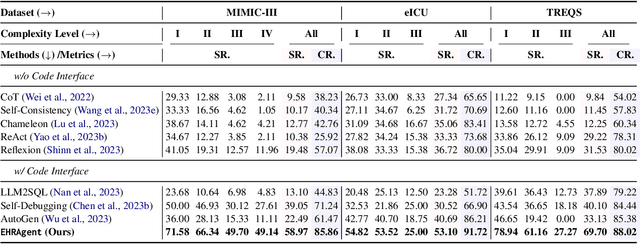
Abstract:Large language models (LLMs) have demonstrated exceptional capabilities in planning and tool utilization as autonomous agents, but few have been developed for medical problem-solving. We propose EHRAgent1, an LLM agent empowered with a code interface, to autonomously generate and execute code for complex clinical tasks within electronic health records (EHRs). First, we formulate an EHR question-answering task into a tool-use planning process, efficiently decomposing a complicated task into a sequence of manageable actions. By integrating interactive coding and execution feedback, EHRAgent learns from error messages and improves the originally generated code through iterations. Furthermore, we enhance the LLM agent by incorporating long-term memory, which allows EHRAgent to effectively select and build upon the most relevant successful cases from past experiences. Experiments on two real-world EHR datasets show that EHRAgent outperforms the strongest LLM agent baseline by 36.48% and 12.41%, respectively. EHRAgent leverages the emerging few-shot learning capabilities of LLMs, enabling autonomous code generation and execution to tackle complex clinical tasks with minimal demonstrations.
Explainable Artificial Intelligence Methods in Combating Pandemics: A Systematic Review
Dec 25, 2021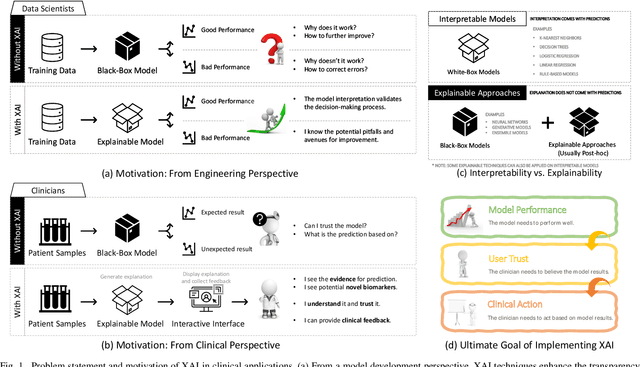
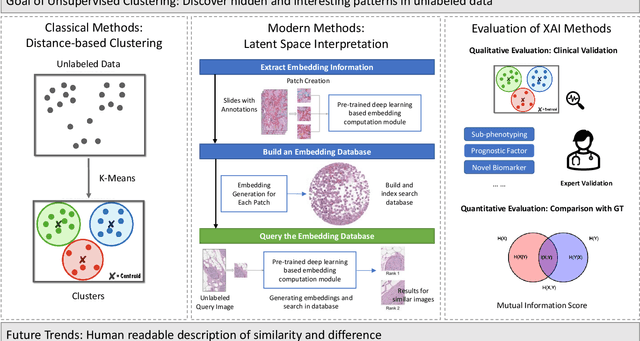
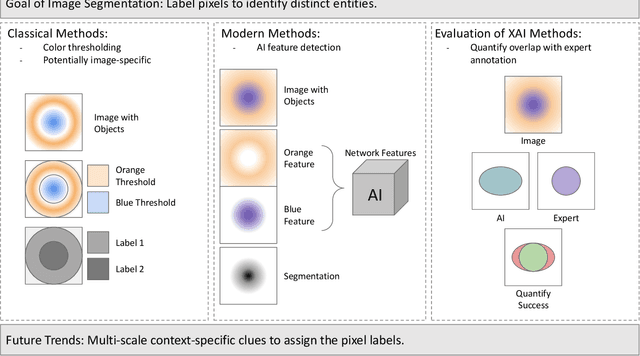
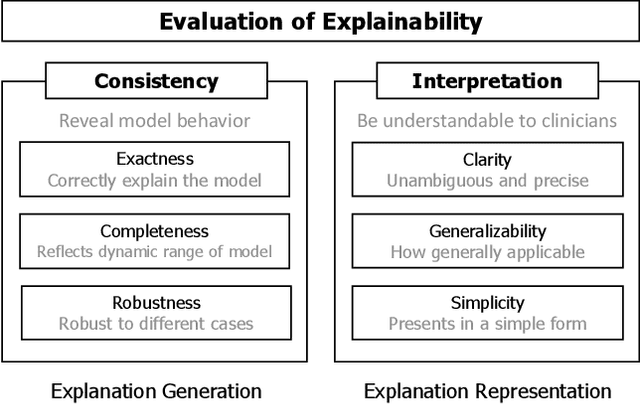
Abstract:Despite the myriad peer-reviewed papers demonstrating novel Artificial Intelligence (AI)-based solutions to COVID-19 challenges during the pandemic, few have made significant clinical impact. The impact of artificial intelligence during the COVID-19 pandemic was greatly limited by lack of model transparency. This systematic review examines the use of Explainable Artificial Intelligence (XAI) during the pandemic and how its use could overcome barriers to real-world success. We find that successful use of XAI can improve model performance, instill trust in the end-user, and provide the value needed to affect user decision-making. We introduce the reader to common XAI techniques, their utility, and specific examples of their application. Evaluation of XAI results is also discussed as an important step to maximize the value of AI-based clinical decision support systems. We illustrate the classical, modern, and potential future trends of XAI to elucidate the evolution of novel XAI techniques. Finally, we provide a checklist of suggestions during the experimental design process supported by recent publications. Common challenges during the implementation of AI solutions are also addressed with specific examples of potential solutions. We hope this review may serve as a guide to improve the clinical impact of future AI-based solutions.
Public Health Informatics: Proposing Causal Sequence of Death Using Neural Machine Translation
Sep 22, 2020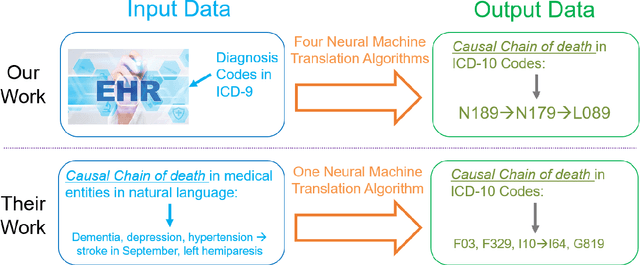
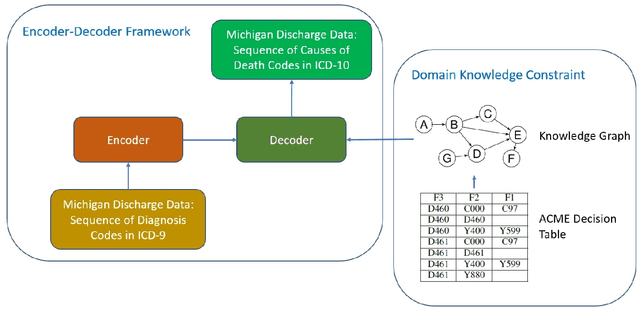
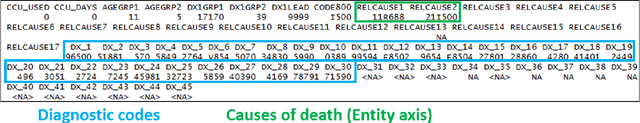
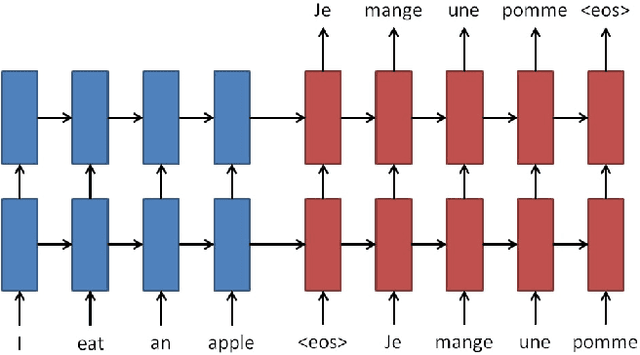
Abstract:Each year there are nearly 57 million deaths around the world, with over 2.7 million in the United States. Timely, accurate and complete death reporting is critical in public health, as institutions and government agencies rely on death reports to analyze vital statistics and to formulate responses to communicable diseases. Inaccurate death reporting may result in potential misdirection of public health policies. Determining the causes of death is, nevertheless, challenging even for experienced physicians. To facilitate physicians in accurately reporting causes of death, we present an advanced AI approach to determine a chronically ordered sequence of clinical conditions that lead to death, based on decedent's last hospital admission discharge record. The sequence of clinical codes on the death report is named as causal chain of death, coded in the tenth revision of International Statistical Classification of Diseases (ICD-10); the priority-ordered clinical conditions on the discharge record are coded in ICD-9. We identify three challenges in proposing the causal chain of death: two versions of coding system in clinical codes, medical domain knowledge conflict, and data interoperability. To overcome the first challenge in this sequence-to-sequence problem, we apply neural machine translation models to generate target sequence. We evaluate the quality of generated sequences with the BLEU (BiLingual Evaluation Understudy) score and achieve 16.44 out of 100. To address the second challenge, we incorporate expert-verified medical domain knowledge as constraint in generating output sequence to exclude infeasible causal chains. Lastly, we demonstrate the usability of our work in a Fast Healthcare Interoperability Resources (FHIR) interface to address the third challenge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge